


The functions estimating tree volume and aboveground (ABG) phytomass for the 2nd Italian National Forest Inventory (2005) are documented in Tabacchi et al. (2011). (The section of the the publication exposing “Methods and results global evaluation” is accessible here Regression models have been developed based on sample trees collected within three successive projects, in order to consider, as much as possible, all of the most relevant tree species present in Italian forests.
Providing such a general and uniform estimation tool, these functions
have been adopted by Italian foresters as reference functions. Many
professionals and researchers have copied, from the paper print or the
PDF file, the small subset of coefficient values required to tackle
their problem at hand.
The package offers an easier (and more reliable) way to proceed.
Moreover, since coefficients were not readily available through a
digital tool, the valuable information that the work developed for the
evaluation of estimates accuracy, has been practically ignored.
Using the ForIT package, all estimates are associated with
accuracy evaluations, through attributes.
The independent variables used in the model are the diameter-at-breast-height (\(`dbh`\)) and the total tree height (\(`h_{tot}`\)). The general model formulation adopted is the following:
{y_0 = b_0 + b_1 × dbh^2 × h_{tot} + b_2 × dbh}where \(`y_0`\) is the volume or AGB phytomass.
Through the ForIT package, all the Tabacchi et
al. (2011) volume and phytomass estimation functions are readily
available in the R environment.
The version under development can be installed using:
devtools::install_gitlab("NuoroForestrySchool/forit") library(tidyverse)
library(ForIT)The package functions cover three main areas:
1) the estimation of tree quantities, for single trees or as
totals;
2) tabulation, to obtain the volume or phytomass tables
(INFCtabulate());
3) graphical accuracy assessment (INFCaccuracyPlot0() and
INFCaccuracyPlot()).
Connecting the package with well established operational standards,
the ‘tree species codes’ used by ForIT are the “EPPO
codes”. (See https://gd.eppo.int, for more info on EPPO project and
data(INFCspecies) for a complete list of species used in
this package).
Single tree estimates are obtained using the INFCvpe()
function.
Otherwise, if interested in estimates for groups of trees, one can refer
to the INFCvpeSUM family of functions.
Estimation of the volume of one Acer campestre tree (EPPO code is ‘ACRCA’) with diameter-at-breast-height equal to 22 cm and 14 m height.
vol <- INFCvpe("ACRCA", dbh.cm = 22, htot.m = 14)The default estimated quantity is volume
(quantity = "vol"). In this specific case, the estimated
volume is 252.96 dm3.
> round(as.numeric(vol),2)
# [1] 252.96The codes and definitions of all the quantities the functions estimate are listed in the ‘Quantities’ table:
> ForIT::Quantities
# # A tibble: 5 × 2
# quantity quantity_definition
# <chr> <chr>
# 1 vol volume of the stem and large branches [dm^3]
# 2 dw1 phytomass of the stem and large braches [kg]
# 3 dw2 phytomass of the small branches [kg]
# 4 dw3 phytomass of the stump [kg]
# 5 dw4 phytomass of the whole tree [kg] Volume is expressed in [dm3], while all the quantities referred to phytomass are expressed in [kg].
The attributes accompanying each estimated value, specify accuracy
evaluations for the selected quantity. wrv is the ;
Var_ea is the variance for an estimated average;
Var_ie the variance for an individual estimate;
InDomain is a logical indicating whether the (dbh, htot)
point lies out of the domain explored by the experimental data (see
INFCtabulate()).
> vol
# [1] 252.9581
# attr(,"pag") # page number, referred to original source
# [1] 231
# attr(,"quantity") # see above
# [1] "vol"
# attr(,"wrv") # weighted residual variance
# [1] 2.271e-05
# attr(,"Var_ea") # variance for an estimated average
# [1] 33.17182
# attr(,"Var_ie") # variance for an individual estimate
# [1] 1075.883
# attr(,"InDomain") # logical indicating whether the (dbh, htot) point lies within the domain
# [1] TRUECumulative estimation of the volume or phytomass of groups of trees is just the summation of the values computed with INFCvpe(), but the computation of accuracy estimates is improved using these summation functions.
Two approaches are available.
INFCvpe_summarise()This function, alternative to dplyr::summarise(),
returns a dataframe (tibble) with unique values combinations in the
grouping column/s, defined via group_by(), plus a fixed set
of columns for estimates and accuracy evaluation.
ForIT_test_data %>%
INFCvpe_summarise("specie", "d130", "h_dendro")
# A tibble: 1 × 6
# Groups: quantity [1]
# quantity n n_out est cihw p
# <chr> <int> <int> <dbl> <dbl> <dbl>
# 1 vol 17 1 10335. 927. 0.95dplyr::summarise()for each set of rows, defined via group_by(), following
aggregation functions are available: * INFCvpe_sum(), that
returns the sum of the estimated quantities; *
INFCvpe_ConfInt(), that returns ‘confidence interval half
width’; * INFCvpe_OutOfDomain(), that returns the number of
‘out of domain’ (dbh, h_tot) pairs included in the summation.
For more details on the functions of this family:
?ForIT::INFCvpeSUM
INFCtabulateVolume and phytomass functions are tabulated in Tabacchi et al. (2011a). Printed numbers serve as reference to verify that coded functions return expected results and, more specifically, empty spaces (or NA) in the printed tables limit function applicability domain. In other words, measurement data used to estimate function coefficient values, cover only the portion of the (dbh, htot) plane where numbers are printed.
> INFCtabulate("ACRCA")
# EPPO code: ACRCA - pag = 231 - quantity = ' vol '
# htot.m
# dbh.cm 5 8 11 14 17 20 23
# 5 6.3 9.1 NA NA NA NA NA
# 10 20.2 31.4 42.5 NA NA NA NA
# 15 NA 68.4 93.5 118.5 NA NA NA
# 20 NA NA 164.9 209.3 253.8 298.3 NA
# 25 NA NA NA 326.2 395.7 465.2 NA
# 30 NA NA NA 468.9 569.0 669.2 769.3
# 35 NA NA NA NA 773.9 910.2 1046.5
# 40 NA NA NA NA NA 1188.3 1366.3
# ---More info at ?ForIT::INFCtabulate.
The tabulation described in §2 covers a limited region
of the dbh by h_tot rectangle.
This region is the “domain” of the reliable estimates, based on the
distribution of the sample trees used to calibrate the functions. The
coefficient of variation (CV = standard_deviation / estimate) is
computed and plotted (as ‘filled contours’) for the whole rectangular
area, the limits of the region of reliable estimates (the “domain”), is
superimposed as a light coloured line. Function output is a
ggplot object that can be used by itself or as a background
on top of which the user can plot his/her data to verify eventual
accuracy or reliability problems.
INFCaccuracyPlot0() - produces, much faster, the plots
at the finest resolution, using pre-calculated values stored in a
specific auxiliary dataframe (see INFC_CVgrid), necessarily
leaving less customization freedom.
INFCaccuracyPlot0("LAXDE") 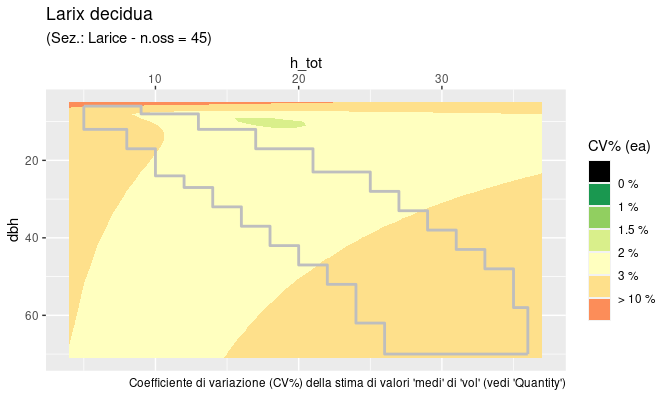
INFCaccuracyPlot() - allows the plots to be fully
customized but, beware, all values required for the ‘fill’ will be
computed and, at finer resolution, the process can be slow.
INFCaccuracyPlot("ABIAL", ie.Var = T, plot.est = T, cv.ul = .15, fixed = F)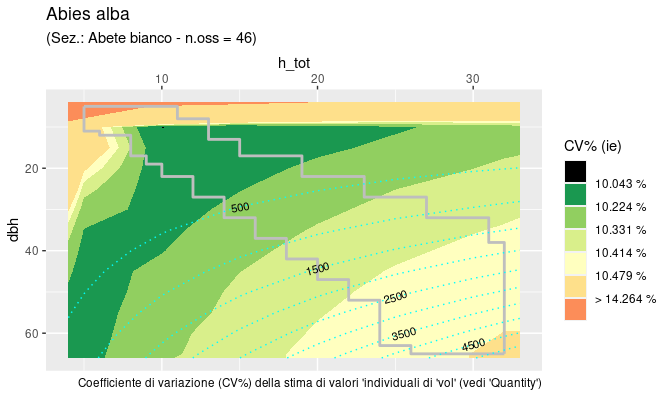
Gasparini, P., Tabacchi, G. (eds), 2011. L’Inventario Nazionale delle Foreste e dei serbatoi forestali di Carbonio INFC 2005. Secondo inventario forestale nazionale italiano. Metodi e risultati. Edagricole. 653 pp.
Tabacchi G., Di Cosmo L., Gasparini P., Morelli S., 2011. Stima del volume e della fitomassa delle principali specie forestali italiane. Equazioni di previsione, tavole del volume e tavole della fitomassa arborea epigea. Stima del volume e della fitomassa delle principali specie forestali italiane. Equazioni di previsione, tavole del volume e tavole della fitomassa arborea epigea. 412 pp. full text
Tabacchi G., Di Cosmo L., Gasparini P., 2011. Aboveground tree volume and phytomass prediction equations for forest species in Italy. European Journal of Forest Research 130: 6 911-934.