This package estimates an ensemble of meta-analytic models (assuming either the presence or absence of effect, heterogeneity, and publication bias) and uses Bayesian model averaging to combine them. The ensemble uses Bayes factors to test for the presence of absence of the individual components (e.g., effect vs. no effect) and model-averages parameter estimates based on posterior model probabilities. The user can define a wide range of non-informative or informative priors for the effect size, heterogeneity, and publication bias components (including selection, PET, and PEESE style models). The package provides convenient functions for summary, visualizations, and fit diagnostics.
See our new pre-print Bartoš et al. (2021) (https://doi.org/10.31234/osf.io/kvsp7) for the description of the newest version, RoBMA-PSMA, or our previous paper introducing the method Maier et al. (in press) (https://doi.org/10.31234/osf.io/u4cns). The previous version of the methods is also implemented within the the user-friendly graphical user interface of JASP (JASP Team, 2020) and accompanied by a tutorial paper with more examples (Bartoš et al., in press) (https://doi.org/10.31234/osf.io/75bqn).
We also prepared multiple vignettes that illustrate functionality of the package:
The package was updated to version 2.0 to provides Bayesian model-averaging across selection models and PET-PEESE (as described in Bartoš et al. (2021) at https://doi.org/10.31234/osf.io/kvsp7).
Please note that the RoBMA 2.0 is not backwards compatible with the previous versions of RoBMA. Use
remotes::install_version("RoBMA", version = "1.2.1")to install the previous version if needed. (Or use the source packages archived with at OSF repositories associated with the corresponding projects.)
The 2.0 version brings several updates to the package:
priors_mu -> priors_effect,
priors_tau -> priors_heterogeneity, and
priors_omega -> priors_bias),prior(distribution = "two.sided", parameters = ...) ->
prior_weightfunction(distribution = "two.sided", parameters = ...)),prior_none()),prior_PET(distribution = "Cauchy", parameters = ...) and
prior_PEESE(distribution = "Cauchy", parameters = ...)),model_type argument allowing to specify different
“pre-canned” models ("PSMA" = RoBMA-PSMA, "PP"
= RoBMA-PP, "2w" = corresponding to Maier et al. (in
press)),combine_data function allows combination of different
effect sizes / variability measures into a common effect size measure
(also used from within the RoBMA function)autofit = FALSE)prior_scale and
transformation arguments),The package requires JAGS 4.3.1 to be installed. The release version can be installed from CRAN:
install.packages("RoBMA")and the development version of the package can be installed from GitHub:
devtools::install_github("fbartos/RoBMA")To illustrate the functionality of the package, we fit the RoBMA-PSMA model from the example in Bartoš et al. (2021) to adjust for publication bias in the infamous Bem (2011) “Feeling the future” pre-cognition study. The RoBMA-PSMA model combines six selection models and PET-PEESE to adjust for publication bias. As in the pre-print, we analyze the data as described by Bem et al. (2011) in his reply to methodological critiques.
First, we load the package and the data set included in the package.
library(RoBMA)
#> Loading required namespace: runjags
#> Loading required namespace: mvtnorm
#> module RoBMA loaded
data("Bem2011", package = "RoBMA")
Bem2011
#> d se study
#> 1 0.25 0.10155048 Detection of Erotic Stimuli
#> 2 0.20 0.08246211 Avoidance of Negative Stimuli
#> 3 0.26 0.10323629 Retroactive Priming I
#> 4 0.23 0.10182427 Retroactive Priming II
#> 5 0.22 0.10120277 Retroactive Habituation I - Negative trials
#> 6 0.15 0.08210765 Retroactive Habituation II - Negative trials
#> 7 0.09 0.07085372 Retroactive Induction of Boredom
#> 8 0.19 0.10089846 Facilitation of Recall I
#> 9 0.42 0.14752627 Facilitation of Recall IIThen, we fit the meta-analytic model ensemble that is composed of 36 models (the new default settings of RoBMA fitting function). These models represent all possible combinations of prior distributions for the following components:
The prior odds of the components are by default set to make all three model categories equally likely a priory (0.5 prior probability of the presence of the effect, 0.5 prior probability of the presence of the heterogeneity, and 0.5 prior probability of the presence of the publication bias). The prior model probability of the publication bias adjustment component is further split equally among the selection models represented by the six weightfunctions and the PET-PEESE models.
fit <- RoBMA(d = Bem2011$d, se = Bem2011$se, study_names = Bem2011$study, seed = 1)The main summary can be obtained using the
summary.RoBMA() function.
The first table shows an overview of the ensemble composition. The
number of models, the prior and posterior model probabilities, and
inclusion Bayes factor of the ensemble components representing the
alternative hypothesis of the presence of the effect, heterogeneity, and
publication bias, We can see the data show very weak evidence, barely
worth mentioning, against the presence of the effect ( ->
), moderate
evidence for the absence of heterogeneity (
->
), and
strong evidence for the presence of publication bias (
).
The second table shows model-averaged estimates weighted by the
individual models’ posterior probabilities. The mean estimate , 95% CI [-0.041, 0.213], is very
close to zero, corresponding to the a priory expected absence of
pre-cognition. The heterogeneity estimate
has most of its probability mass around zero
due to the higher support of models assuming absence of the
heterogeneity. The parameters omega, representing the publication
weights at each p-value interval are decreasing with increasing
p-values, showing the publication bias, as well as the non zero
PET and PEESE estimates.
summary(fit)
#> Call:
#> RoBMA(d = Bem2011$d, se = Bem2011$se, study_names = Bem2011$study,
#> seed = 1)
#>
#> Robust Bayesian meta-analysis
#> Components summary:
#> Models Prior prob. Post. prob. Inclusion BF
#> Effect 18/36 0.500 0.324 0.479
#> Heterogeneity 18/36 0.500 0.125 0.143
#> Bias 32/36 0.500 0.942 16.323
#>
#> Model-averaged estimates:
#> Mean Median 0.025 0.975
#> mu 0.037 0.000 -0.041 0.213
#> tau 0.010 0.000 0.000 0.113
#> omega[0,0.025] 1.000 1.000 1.000 1.000
#> omega[0.025,0.05] 0.935 1.000 0.338 1.000
#> omega[0.05,0.5] 0.780 1.000 0.009 1.000
#> omega[0.5,0.95] 0.768 1.000 0.007 1.000
#> omega[0.95,0.975] 0.786 1.000 0.007 1.000
#> omega[0.975,1] 0.801 1.000 0.007 1.000
#> PET 0.759 0.000 0.000 2.805
#> PEESE 6.183 0.000 0.000 25.463
#> The estimates are summarized on the Cohen's d scale (priors were specified on the Cohen's d scale).
#> (Estimated publication weights omega correspond to one-sided p-values.)We can visualize the estimated mean and heterogeneity parameters
using the plot.RoBMA() function. The arrows in both figures
represent the point probability mass at and
, corresponding to the null hypotheses
of the absence of effect and heterogeneity, both increasing in the
posterior model probability from 0.5 to 0.676 and 0.875
respectively.
plot(fit, parameter = "mu", xlim = c(-0.5, 0.5))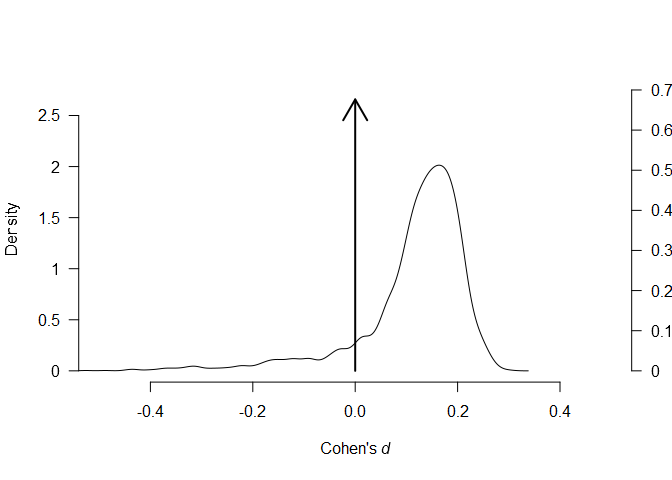
plot(fit, parameter = "tau")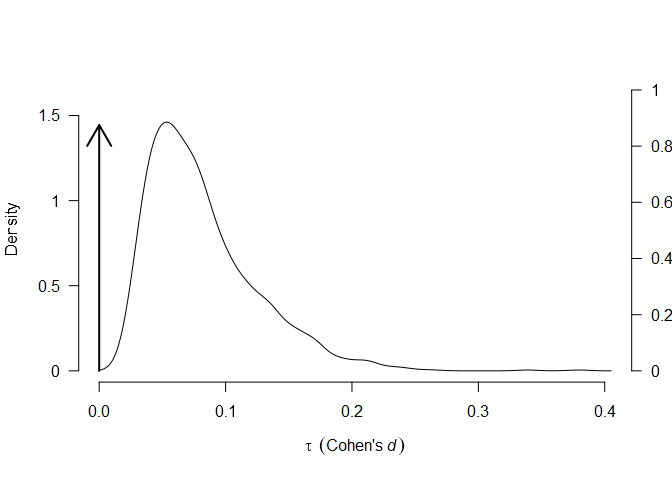
We can further visualize the publication bias adjustments of selection models, visualizing the posterior estimate of the model-averaged weightfunction that shows a sharp decrease in the publication weights of studies with p-values above the “marginal significance” (0.10) level,
plot(fit, parameter = "weightfunction", rescale_x = TRUE)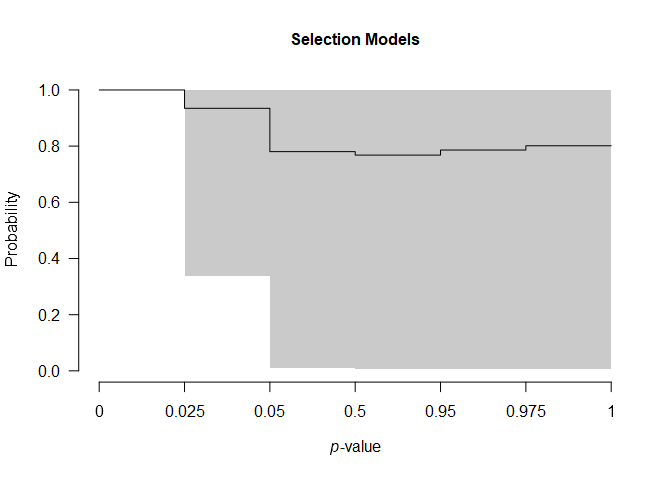
and the PET-PEESE publication bias adjustment, visualizing the individual studies’ standard errors and effect sizes as diamonds and the model-averaged estimate of the regression lines that shows a steady increase of effect sizes with increasing standard errors.
plot(fit, parameter = "PET-PEESE", xlim = c(0, 0.25))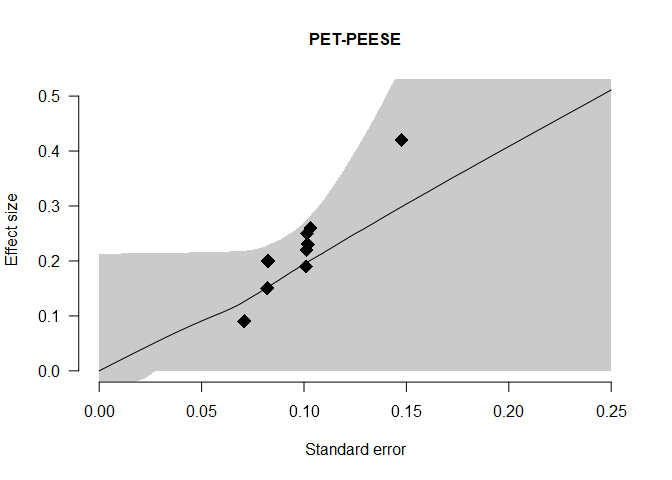
The usual meta-analytic forest plot can be obtained with the
forest() function,
forest(fit)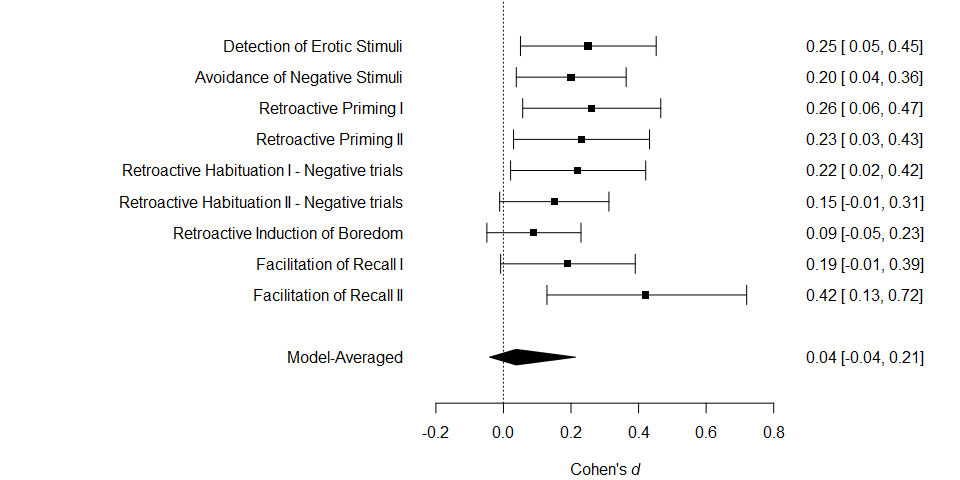
and visualization of the effect size estimates from models assuming
presence of the effect can be obtained with the
plot_models() function.
plot_models(fit, conditional = TRUE)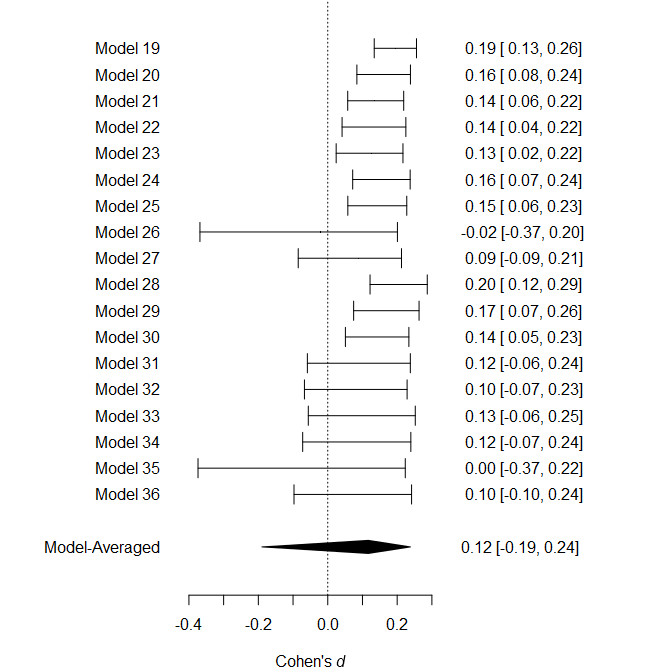
Apart from plotting, the individual model performance can be
inspected using the summary.RoBMA() function with argument
type = "models" or the overview of the individual model
MCMC diagnostics can be obtained by setting
type = "diagnostics" (not shown here for the lack of
space).
We can also visualize the MCMC diagnostics using the diagnostics
function. The function can display the chains
type = "chain" / posterior sample densities
type = "densities", and averaged auto-correlations
type = "autocorrelation". Here, we request the chains trace
plot of the parameter of the most complex model by setting
show_models = 36 (the model numbers can be obtained from
the summary function with type = "models" argument).
diagnostics(fit, parameter = "mu", type = "chains", show_models = 36)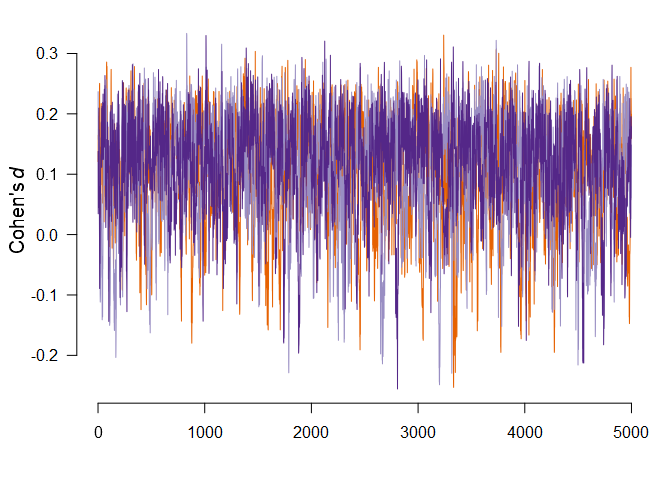
The package allows to fit highly customized models with different
prior distribution functions, prior model probabilities, and provides
more visualization options. See the documentation to find out more about
the specific functions: RoBMA(), priors(),
plot.RoBMA(). The main package functionality is also
implemented within the Meta Analysis module of JASP 0.14 (JASP Team,
2020) and will be soon updated to accommodate the 2.0 version of the
package.