


Welcome to cobalt, which stands for
Covariate Balance
Tables (and Plots). cobalt allows users to
assess balance on covariate distributions in preprocessed groups
generated through weighting, matching, or subclassification, such as by
using the propensity score. cobalt’s primary function is
bal.tab(), which stands for “balance table”, and is meant
to replace (or supplement) the balance assessment tools found in other R
packages. To examine how bal.tab() integrates with these
packages and others, see the help file for bal.tab() with
?bal.tab, which links to the methods used for each package.
Each page has examples of how bal.tab() is used with the
package. There are also five vignettes detailing the use of
cobalt, which can be accessed at
vignette("cobalt"): one for basic uses of
cobalt, one for the use of cobalt with
additional packages, one for the use of cobalt with
multiply imputed and/or clustered data, one for the use of
cobalt with longitudinal treatments, and one for the use of
cobalt to generate publication-ready plots. Currently,
cobalt is compatible with output from MatchIt,
twang, Matching, optmatch,
CBPS, ebal, WeightIt,
designmatch, sbw, MatchThem, and
cem as well as data not processed through these
packages.
Most of the major conditioning packages contain functions to assess
balance; so why use cobalt at all? cobalt
arose out of several desiderata when using these packages: to have
standardized measures that were consistent across all conditioning
packages, to allow for flexibility in the calculation and display of
balance measures, and to incorporate recent methodological
recommendations in the assessment of balance. In addition,
cobalt has unique plotting capabilities that make use of
ggplot2 in R for balance assessment and reporting.
Because conditioning methods are spread across several packages which
each have their idiosyncrasies in how they report balance (if at all),
comparing the resulting balance from various conditioning methods can be
a challenge. cobalt unites these packages by providing a
single, flexible tool that intelligently processes output from any of
the conditioning packages and provides the user with both useful
defaults and customizable options for display and calculation.
cobalt also allows for balance assessment on data not
generated through any of the conditioning packages. In addition,
cobalt has tools for assessing and reporting balance for
clustered data sets, data sets generated through multiple imputation,
and data sets with a continuous treatment variable, all features that
exist in very limited capacities or not at all in other packages.
A large focus in developing cobalt was to streamline
output so that only the most useful, non-redundant, and complete
information is displayed, all at the user’s choice. Balance statistics
are intuitive, methodologically informed, and simple to interpret.
Visual displays of balance reflect the goals of balance assessment
rather than being steps removed. While other packages have focused their
efforts on processing data, cobalt only assesses balance,
and does so particularly well.
New features are being added all the time, following the cutting edge
of methodological work on balance assessment. As new packages and
methods are developed, cobalt will be ready to integrate
them to further our goal of simple, unified balance assessment.
Below are examples of cobalt’s primary functions:
library("cobalt")
data("lalonde", package = "cobalt")
# Nearest neighbor matching with MatchIt
m.out <- MatchIt::matchit(treat ~ age + educ + race + married + nodegree + re74 +
re75, data = lalonde)
# Checking balance before and after matching:
bal.tab(m.out, thresholds = c(m = 0.1), un = TRUE)#> Call
#> MatchIt::matchit(formula = treat ~ age + educ + race + married +
#> nodegree + re74 + re75, data = lalonde)
#>
#> Balance Measures
#> Type Diff.Un Diff.Adj M.Threshold
#> distance Distance 1.7941 0.9739
#> age Contin. -0.3094 0.0718 Balanced, <0.1
#> educ Contin. 0.0550 -0.1290 Not Balanced, >0.1
#> race_black Binary 0.6404 0.3730 Not Balanced, >0.1
#> race_hispan Binary -0.0827 -0.1568 Not Balanced, >0.1
#> race_white Binary -0.5577 -0.2162 Not Balanced, >0.1
#> married Binary -0.3236 -0.0216 Balanced, <0.1
#> nodegree Binary 0.1114 0.0703 Balanced, <0.1
#> re74 Contin. -0.7211 -0.0505 Balanced, <0.1
#> re75 Contin. -0.2903 -0.0257 Balanced, <0.1
#>
#> Balance tally for mean differences
#> count
#> Balanced, <0.1 5
#> Not Balanced, >0.1 4
#>
#> Variable with the greatest mean difference
#> Variable Diff.Adj M.Threshold
#> race_black 0.373 Not Balanced, >0.1
#>
#> Sample sizes
#> Control Treated
#> All 429 185
#> Matched 185 185
#> Unmatched 244 0# Examining distributional balance with plots:
bal.plot(m.out, var.name = "educ")
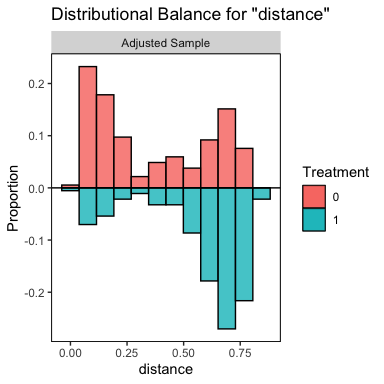
bal.plot(m.out, var.name = "distance", mirror = TRUE, type = "histogram")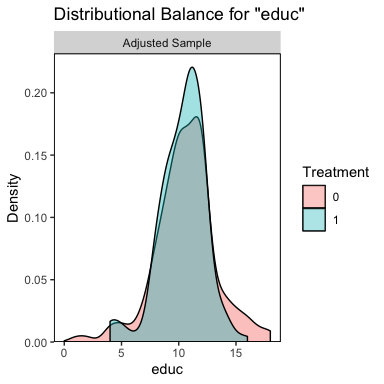
# Generating a Love plot to report balance:
love.plot(m.out, stats = c("mean.diffs", "variance.ratios"), thresholds = c(m = 0.1,
v = 2), abs = TRUE, binary = "std", var.order = "unadjusted")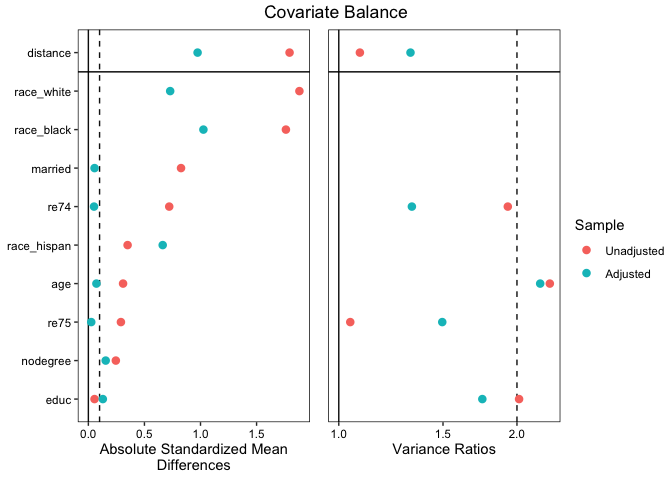
Please remember to cite this package when using it to analyze data.
For example, in a manuscript, you could write: “Matching was performed
using the Matching package (Sekhon, 2011), and covariate
balance was assessed using cobalt (Greifer, 2022), both in R (R
Core Team, 2022).” Use citation("cobalt") to generate a
bibliographic reference for the cobalt package.
Bugs appear in cobalt occasionally, often found by
users. Please report any bugs at https://github.com/ngreifer/cobalt/issues. To install
the latest development version of cobalt, which may have
removed a bug you’re experiencing, use the following code:
devtools::install_github("ngreifer/cobalt")