cytofan implements the concept of fan plots (Britton, E.; Fisher, P. & J. Whitley (1998) The Inflation Report Projections: Understanding the Fan Chart) for cytometry data in ggplot2.
The cytofan package was implemented following up on a gist written shortly after the fanplot package was released. Compared to ggfan, cytofan uses categorical data as input on the x axis.
You can install cytofan from github with:
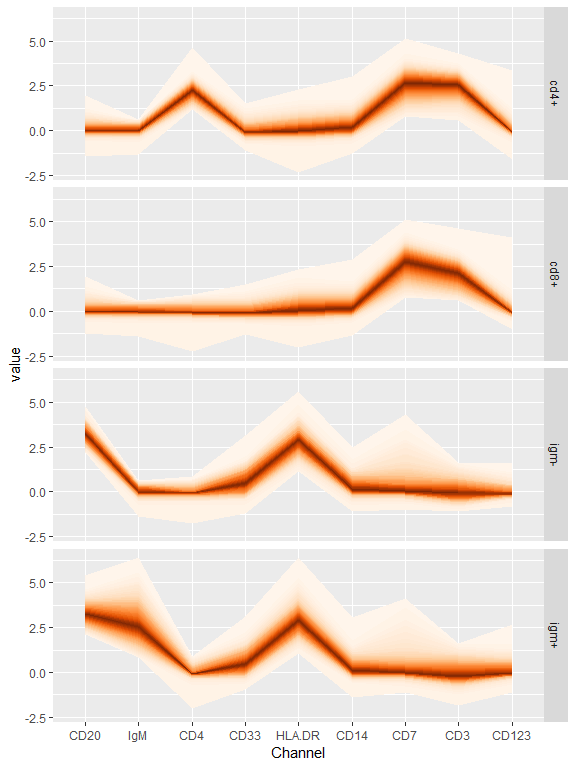
cytofan can be used to visualize differences between populations identifed using mass cytometry:
library(cytofan)
#> Loading required package: ggplot2
library(bodenmiller)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(reshape2)
data("refPhenoMat")
data("refAnnots")
bind_cols(refAnnots,
as.data.frame(refPhenoMat)) %>%
melt(.,measure.vars=colnames(refPhenoMat),
variable.name='Channel') %>%
filter(Cells %in% c('cd4+','cd8+','igm+','igm-')) %>%
ggplot(aes(x=Channel,y=value))+
geom_fan()+
facet_grid(Cells~.)