 driveR: An R Package for Prioritizing Cancer Driver Genes Using Genomics
Data
driveR: An R Package for Prioritizing Cancer Driver Genes Using Genomics
Data driveR: An R Package for Prioritizing Cancer Driver Genes Using Genomics
Data
driveR: An R Package for Prioritizing Cancer Driver Genes Using Genomics
DataCancer genomes contain large numbers of somatic alterations but few genes drive tumor development. Identifying cancer driver genes is critical for precision oncology. Most of current approaches either identify driver genes based on mutational recurrence or using estimated scores predicting the functional consequences of mutations.
driveR is a tool for personalized or batch analysis of
genomics data for driver gene prioritization by combining genomics
information and prior biological knowledge. As features, driveR uses
coding impact metaprediction scores, non-coding impact scores, somatic
copy number alteration scores, hotspot gene/double-hit gene condition,
‘phenolyzer’ gene scores and memberships to cancer-related KEGG
pathways. It uses these features to estimate cancer-type-specific
probabilities for each gene of being a cancer driver using the related
task of a multi-task learning classification model.
The method is described in detail in Ülgen E, Sezerman OU. driveR: a novel method for prioritizing cancer driver genes using somatic genomics data. BMC Bioinformatics. 2021 May 24;22(1):263.https://doi.org/10.1186/s12859-021-04203-7
You can install the latest released version of driveR
from CRAN via:
install.packages("driveR")You can install the development version of driveR from
GitHub with:
# install.packages("devtools")
devtools::install_github("egeulgen/driveR", build_vignettes = TRUE)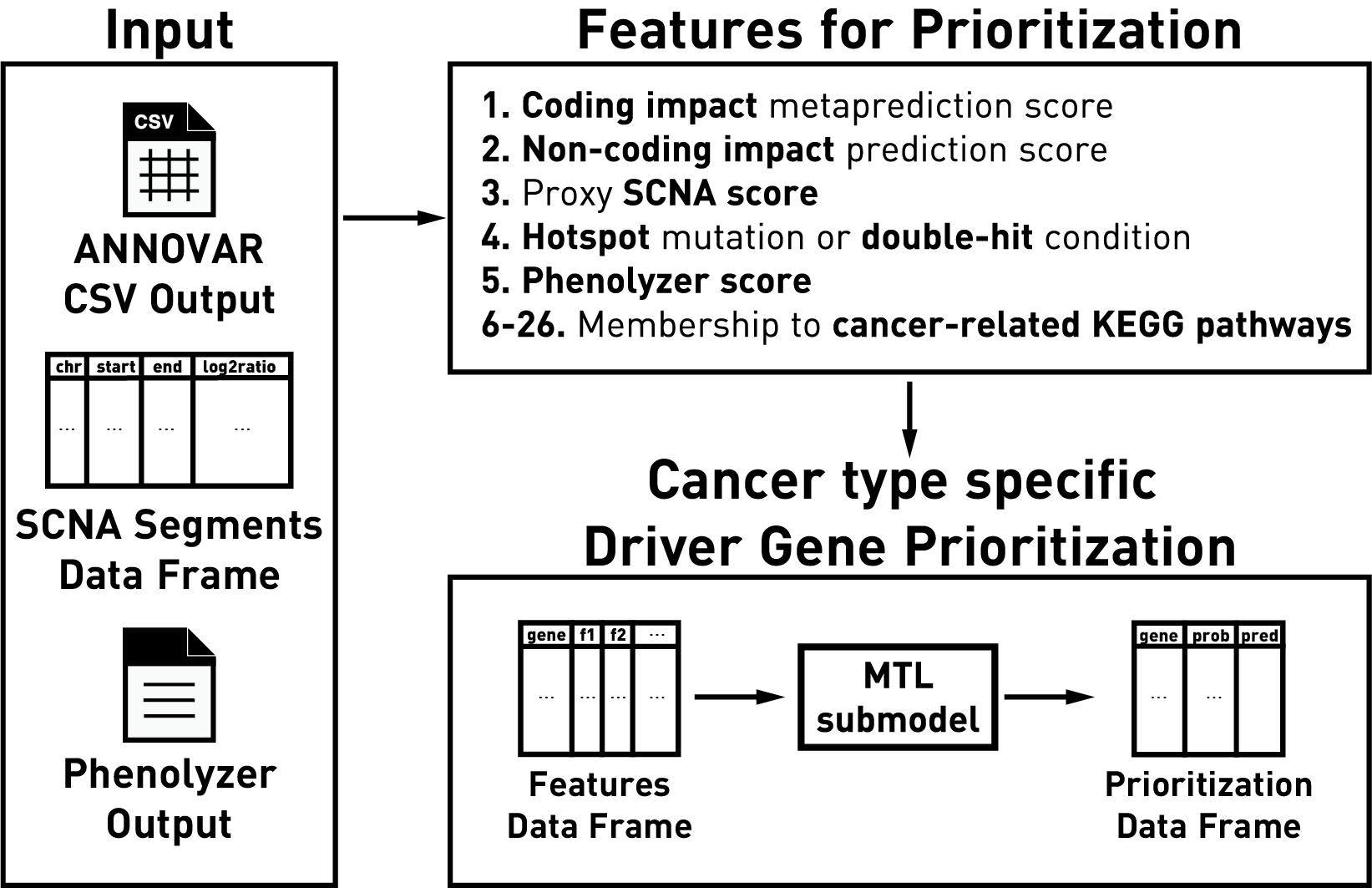
driveR has two main objectives:
predict_coding_impact())create_features_df() and
prioritize_driver_genes())Note that driveR require operations outside of R and
depends on the outputs from the external tools ANNOVAR and
phenolyzer.
For detailed information on how to use driveR, please
see the vignette “How to use driveR” via
vignette("how_to_use")