
foieGras - fit latent variable movement models to animal tracking data for location quality control and behavioural inference
master branch:
dev branch:
foieGras is an R package that fits a continuous-time model (RW or CRW) in state-space form to filter Argos (or GLS) satellite location data. Template Model Builder (TMB) is used for fast estimation. Argos data can be either (older) Least Squares-based locations, (newer) Kalman Filter-based locations with error ellipse information, or a mixture of the two. The state-space model estimates two sets of location states: 1) corresponding to each observation, which are usually irregularly timed (fitted states); and 2) corresponding to (usually) regular time intervals specified by the user (predicted states). Locations are returned as both LongLat and on the Mercator projection (units=km). Additional models are provided to infer movement behaviour along the SSM-estimated most-probable track.
First, ensure you have R version >= 3.6.0 installed (preferably R 4.0.0 or higher):
foieGras is on CRAN and can be downloaded within R, in the usual way install.packages("foieGras") or, more completely: install.packages("foieGras", depedencies = c("Imports","LinkingTo","Suggests"))
On PC’s running Windows, ensure you have installed Rtools
On Mac’s, ensure you have installed the Command Line Tools for Xcode by executing xcode-select --install in the terminal; or you can download the latest version from the URL (free developer registration may be required). A full Xcode install uses up a lot of disk space and is not required.
To get the very latest foieGras stable version, you can install from GitHub:
Note: there can be issues getting compilers to work properly, especially on a Mac with OS X 10.13.x or higher. If you encounter install and compile issues, I recommend you consult the excellent information on the glmmTMB GitHub.
foieGras is intended to be as easy to use as possible. Here’s an example showing how to quality-control Argos tracking data, and infer a behavioural index along the estimated animal tracks:
library(tidyverse)
library(foieGras)
fit <- fit_ssm(sese, vmax= 4, model = "crw", time.step = 24, control = ssm_control(verbose = 0, se = FALSE))
fmp <- fit_mpm(fit, what = "predicted", model = "jmpm", control = mpm_control(verbose = 0))
plot(fmp, pages = 1, ncol = 3, pal = "Zissou1", rev = TRUE)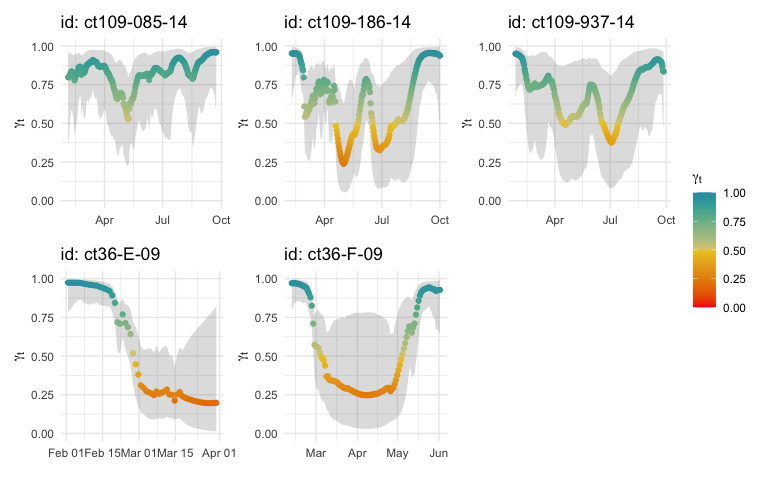
If you are convinced you have encountered a bug or unexpected/inconsistent behaviour when using foieGras, you can post an issue here. First, have a read through the posted issues to see if others have encountered the same problem and whether a solution has been offered. You can reply to an existing issue if you have the same problem and have more details to share or you can submit a new issue. To submit an issue, you will need to clearly describe the unexpected behaviour, include a reproducible example with a small dataset, clearly describe what you expected to happen (but didn’t), and (ideally) post a few screenshots/images that nicely illustrate the problem.
Contributions from anyone in the Movement Ecology/Bio-Logging communities are welcome. Consider submitting a feature request here to start a discussion. Alternatively, if your idea is well-developed then you can submit a pull request for evaluation here. Unsure about what all this means but still want to discuss your idea? then have a look through the GitHub pages of community-built R packages like tidyverse/dplyr for examples.
Please note that the foieGras project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.