

Package ‘ggpmisc’ (Miscellaneous Extensions to ‘ggplot2’) is a set of extensions to R package ‘ggplot2’ (>= 3.0.0) with emphasis on annotations and plotting related to fitted models. Estimates from model fit objects can be displayed in ggplots as text, tables or equations. Predicted values, residuals, deviations and weights can be plotted for various model fit functions. Linear models, quantile regression and major axis regression as well as those functions with accessors following the syntax of package ‘broom’ are supported. Package ‘ggpmisc’ continues to give access to extensions moved as of version 0.4.0 to package ‘ggpp’.
Package ‘ggpmisc’ is consistent with the grammar of graphics, and opens new possibilities retaining the flexibility inherent to this grammar. Its aim is not to automate plotting or annotations in a way suitable for fast data exploration by use of a “fits-all-sizes” predefined design. Package ‘ggpmisc’ together with package ‘ggpp’, provide new layer functions, position functions and scales. In fact, these packages follow the tenets of the grammar even more strictly than ‘ggplot2’ in the distinction between geometries and statistics. The new statistics in ‘ggpmisc’ focus mainly on model fitting, but there is not yet support for multiple comparisons among groups. The default annotations are those most broadly valid and of easiest interpretation. We follow R’s approach of expecting that users know what they need or want, and will usually want to adjust how results from model fits are presented both graphically and textually. The approach and mechanics of plot construction and rendering remain unchanged from those implemented in package ‘ggplot2’.
Statistics that help with reporting the results of model fits are:
| Statistic | Returned values (default geometry) | Met | hods |
|---|---|---|
stat_poly_eq() |
equation, R2, P, etc.
(text_npc) |
lm, rlm (1, 2, 7) |
stat_ma_eq() |
equation, R2, P, etc.
(text_npc) |
lmodel2 (6, 7) |
stat_quant_eq() |
equation, P, etc. (text_npc) |
rq (1, 3, 4, 7) |
stat_correlation() |
correlation, P-value, CI (text_npc) |
Pearson (t), Kendall (z), Spearman (S) |
stat_poly_line() |
line + conf. (smooth) |
lm, rlm (1, 2, 7) |
stat_ma_line() |
line + conf. (smooth) |
lmodel2 (6, 7) |
stat_quant_line() |
line + conf. (smooth) |
rq, rqss (1, 3, 4, 7) |
stat_quant_band() |
median + quartiles (smooth) |
rq, rqss (1, 4, 5, 7) |
stat_fit_residuals() |
residuals (point) |
lm, rlm, rq (1, 2, 4, 7, 8) |
stat_fit_deviations() |
deviations from observations (segment) |
lm, rlm, lqs, rq (1, 2, 4, 7, 9) |
stat_fit_glance() |
equation, R2, P, etc.
(text_npc) |
all those supported by ‘broom’ |
stat_fit_augment() |
predicted and other values (smooth) |
all those supported by ‘broom’ |
stat_fit_tidy() |
fit results, e.g., for equation (text_npc) |
all those supported by ‘broom’ |
stat_fit_tb() |
ANOVA and summary tables (table_npc) |
all those supported by ‘broom’ |
Notes: (1) weight aesthetic supported; (2) user defined fit
functions that return an object of a class derived from lm
are supported even if they override the statistic’s formula
argument; (3) unlimited quantiles supported; (4) user defined fit
functions that return an object of a class derived from rq
or rqs are supported even if they override the statistic’s
formula and/or quantiles argument; (5) two and three
quantiles supported; (6) user defined fit functions that return an
object of a class derived from lmodel2 are supported; (7)
method arguments support colon based notation; (8) various
functions if method residuals() defined for returned value;
(9) various functions if method fitted() defined for
returned value.
Statistics stat_peaks() and stat_valleys()
can be used to highlight and/or label maxima and minima in a plot.
Scales scale_x_logFC() and scale_y_logFC()
are suitable for plotting of log fold change data. Scales
scale_x_Pvalue(), scale_y_Pvalue(),
scale_x_FDR() and scale_y_FDR() are suitable
for plotting p-values and adjusted p-values or false
discovery rate (FDR). Default arguments are suitable for volcano and
quadrant plots as used for transcriptomics, metabolomics and similar
data.
Scales scale_colour_outcome(),
scale_fill_outcome() and scale_shape_outcome()
and functions outome2factor(),
threshold2factor(), xy_outcomes2factor() and
xy_thresholds2factor() used together make it easy to map
ternary numeric outputs and logical binary outcomes to color, fill and
shape aesthetics. Default arguments are suitable for volcano, quadrant
and other plots as used for genomics, metabolomics and similar data.
Several geoms and other extensions formerly included in package
‘ggpmisc’ until version 0.3.9 were migrated to package ‘ggpp’. They are
still available when ‘ggpmisc’ is loaded, but the documentation now
resides in the new package ‘ggpp’.
Functions for the manipulation of layers in ggplot objects, together
with statistics and geometries useful for debugging extensions to
package ‘ggplot2’, included in package ‘ggpmisc’ until version 0.2.17
are now in package ‘gginnards’.
library(ggpmisc)
library(ggrepel)
library(broom)In the first example we plot a time series using the specialized
version of ggplot() that converts the time series into a
tibble and maps the x and y aesthetics
automatically. We also highlight and label the peaks using
stat_peaks.
ggplot(lynx, as.numeric = FALSE) + geom_line() +
stat_peaks(colour = "red") +
stat_peaks(geom = "text", colour = "red", angle = 66,
hjust = -0.1, x.label.fmt = "%Y") +
stat_peaks(geom = "rug", colour = "red", sides = "b") +
expand_limits(y = 8000)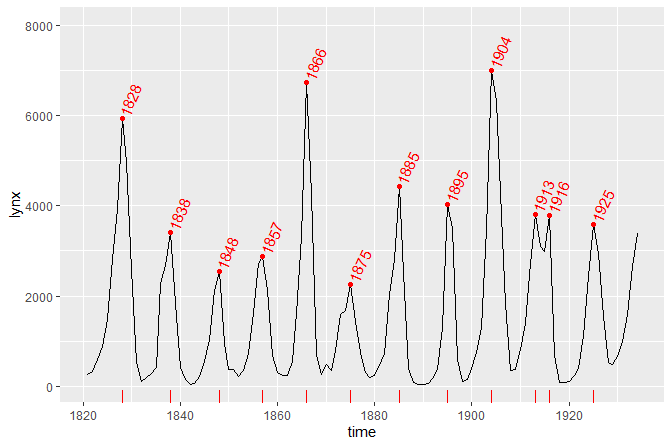
In the second example we add the equation for a fitted polynomial
plus the adjusted coefficient of determination to a plot showing the
observations plus the fitted curve, deviations and confidence band. We
use stat_poly_eq() together with use_label()
to assemble and map the desired annotations.
formula <- y ~ x + I(x^2)
ggplot(cars, aes(speed, dist)) +
geom_point() +
stat_fit_deviations(formula = formula, colour = "red") +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label(c("eq", "adj.R2")), formula = formula)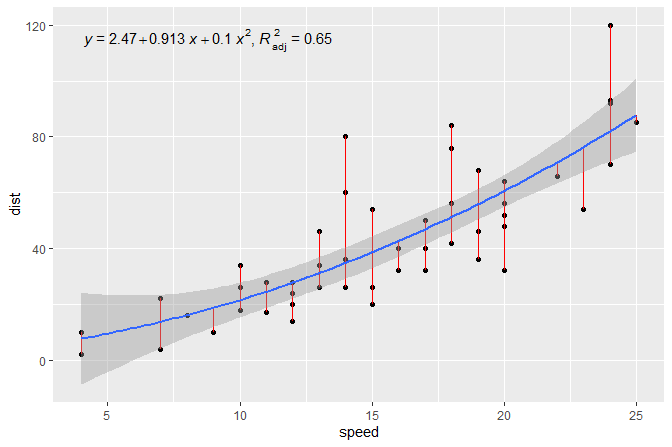
The same figure as in the second example but this time annotated with
the ANOVA table for the model fit. We use stat_fit_tb()
which can be used to add ANOVA or summary tables.
formula <- y ~ x + I(x^2)
ggplot(cars, aes(speed, dist)) +
geom_point() +
geom_smooth(method = "lm", formula = formula) +
stat_fit_tb(method = "lm",
method.args = list(formula = formula),
tb.type = "fit.anova",
tb.vars = c(Effect = "term",
"df",
"M.S." = "meansq",
"italic(F)" = "statistic",
"italic(P)" = "p.value"),
tb.params = c(x = 1, "x^2" = 2),
label.y.npc = "top", label.x.npc = "left",
size = 2.5,
parse = TRUE)
#> Dropping params/terms (rows) from table!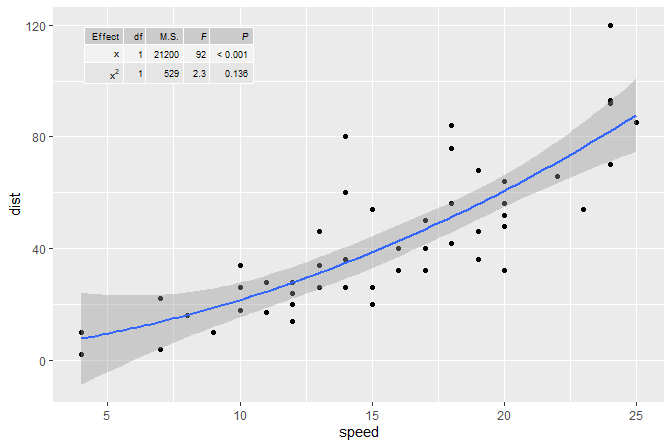
The same figure as in the second example but this time using quantile regression, median in this example.
formula <- y ~ x + I(x^2)
ggplot(cars, aes(speed, dist)) +
geom_point() +
stat_quant_line(formula = formula, quantiles = 0.5) +
stat_quant_eq(formula = formula, quantiles = 0.5)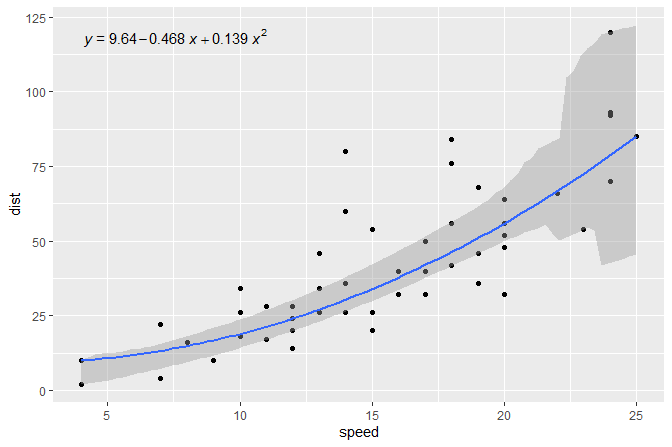
Band highlighting the region between both quartile regressions and a line for the median regression.
formula <- y ~ x + I(x^2)
ggplot(cars, aes(speed, dist)) +
geom_point() +
stat_quant_band(formula = formula)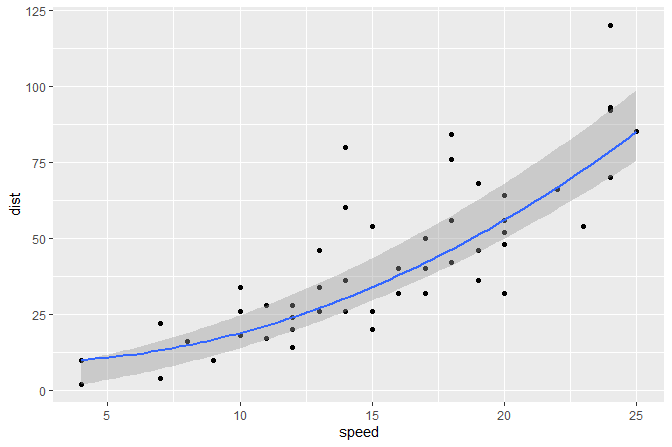
A quadrant plot with counts and labels, using
geom_text_repel() from package ‘ggrepel’.
ggplot(quadrant_example.df, aes(logFC.x, logFC.y)) +
geom_point(alpha = 0.3) +
geom_quadrant_lines() +
stat_quadrant_counts() +
stat_dens2d_filter(color = "red", keep.fraction = 0.02) +
stat_dens2d_labels(aes(label = gene), keep.fraction = 0.02,
geom = "text_repel", size = 2, colour = "red") +
scale_x_logFC(name = "Transcript abundance after A%unit") +
scale_y_logFC(name = "Transcript abundance after B%unit")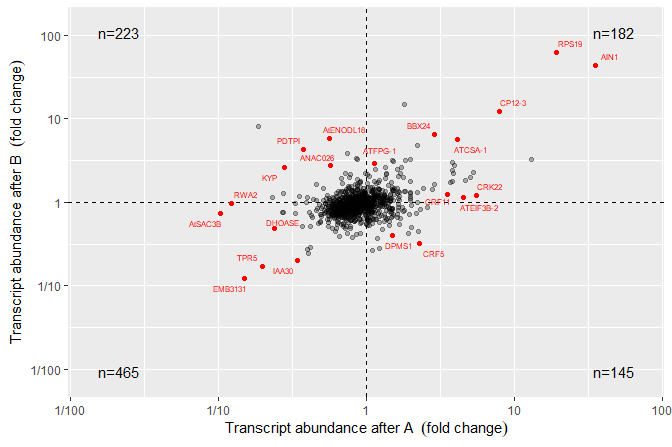
Installation of the most recent stable version from CRAN:
install.packages("ggpmisc")Installation of the current unstable version from GitHub:
# install.packages("devtools")
devtools::install_github("aphalo/ggpmisc")HTML documentation for the package, including help pages and the User Guide, is available at https://docs.r4photobiology.info/ggpmisc/.
News about updates are regularly posted at https://www.r4photobiology.info/.
Chapter 7 in Aphalo (2020) explains basic concepts of the grammar of graphics as implemented in ‘ggplot2’ as well as extensions to this grammar including several of those made available by packages ‘ggpp’ and ‘ggpmisc’. Open access supplementary chapters and other information related to the book is available at https://www.learnr-book.info/.
Please report bugs and request new features at https://github.com/aphalo/ggpmisc/issues. Pull requests are welcome at https://github.com/aphalo/ggpmisc.
If you use this package to produce scientific or commercial publications, please cite according to:
citation("ggpmisc")
#>
#> To cite package 'ggpmisc' in publications use:
#>
#> Aphalo P (2022). _ggpmisc: Miscellaneous Extensions to 'ggplot2'_.
#> https://docs.r4photobiology.info/ggpmisc/,
#> https://github.com/aphalo/ggpmisc.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {ggpmisc: Miscellaneous Extensions to 'ggplot2'},
#> author = {Pedro J. Aphalo},
#> year = {2022},
#> note = {https://docs.r4photobiology.info/ggpmisc/,
#> https://github.com/aphalo/ggpmisc},
#> }Aphalo, Pedro J. (2020) Learn R: As a Language. The R Series. Boca Raton and London: Chapman and Hall/CRC Press. ISBN: 978-0-367-18253-3. 350 pp.
© 2016-2022 Pedro J. Aphalo (pedro.aphalo@helsinki.fi). Released under the GPL, version 2 or greater. This software carries no warranty of any kind.