
{ggstatsplot}:
{ggplot2} Based Plots with Statistical Details| Status | Usage | Miscellaneous |
|---|---|---|
 |

“What is to be sought in designs for the display of information is the clear portrayal of complexity. Not the complication of the simple; rather … the revelation of the complex.” - Edward R. Tufte
{ggstatsplot}
is an extension of {ggplot2}
package for creating graphics with details from statistical tests
included in the information-rich plots themselves. In a typical
exploratory data analysis workflow, data visualization and statistical
modeling are two different phases: visualization informs modeling, and
modeling in its turn can suggest a different visualization method, and
so on and so forth. The central idea of {ggstatsplot} is
simple: combine these two phases into one in the form of graphics with
statistical details, which makes data exploration simpler and
faster.
| Type | Source | Command |
|---|---|---|
| Release | install.packages("ggstatsplot") |
|
| Development | remotes::install_github("IndrajeetPatil/ggstatsplot") |
If you want to cite this package in a scientific journal or in any
other context, run the following code in your R
console:
citation("ggstatsplot")
To cite package 'ggstatsplot' in publications use:
Patil, I. (2021). Visualizations with statistical details: The
'ggstatsplot' approach. Journal of Open Source Software, 6(61), 3167,
doi:10.21105/joss.03167
A BibTeX entry for LaTeX users is
@Article{,
doi = {10.21105/joss.03167},
url = {https://doi.org/10.21105/joss.03167},
year = {2021},
publisher = {{The Open Journal}},
volume = {6},
number = {61},
pages = {3167},
author = {Indrajeet Patil},
title = {{Visualizations with statistical details: The {'ggstatsplot'} approach}},
journal = {{Journal of Open Source Software}},
}I would like to thank all the contributors to
{ggstatsplot} who pointed out bugs or requested features I
hadn’t considered. I would especially like to thank other package
developers (especially Daniel Lüdecke, Dominique Makowski, Mattan S.
Ben-Shachar, Brenton Wiernik, Patrick Mair, Salvatore Mangiafico, etc.)
who have patiently and diligently answered my relentless questions and
supported feature requests in their projects. I also want to thank Chuck
Powell for his initial contributions to the package.
The hexsticker was generously designed by Sarah Otterstetter (Max
Planck Institute for Human Development, Berlin). This package has also
benefited from the larger #rstats community on Twitter,
LinkedIn, and StackOverflow.
Thanks are also due to my postdoc advisers (Mina Cikara and Fiery Cushman at Harvard University; Iyad Rahwan at Max Planck Institute for Human Development) who patiently supported me spending hundreds (?) of hours working on this package rather than what I was paid to do. 😁
To see the detailed documentation for each function in the stable CRAN version of the package, see:
It, therefore, produces a limited kinds of plots for the supported analyses:
In addition to these basic plots, {ggstatsplot} also
provides grouped_ versions (see below)
that makes it easy to repeat the same analysis for any grouping
variable.
The table below summarizes all the different types of analyses currently supported in this package-
| Functions | Description | Parametric | Non-parametric | Robust | Bayesian |
|---|---|---|---|---|---|
ggbetweenstats |
Between group/condition comparisons | ✅ | ✅ | ✅ | ✅ |
ggwithinstats |
Within group/condition comparisons | ✅ | ✅ | ✅ | ✅ |
gghistostats, ggdotplotstats |
Distribution of a numeric variable | ✅ | ✅ | ✅ | ✅ |
ggcorrmat |
Correlation matrix | ✅ | ✅ | ✅ | ✅ |
ggscatterstats |
Correlation between two variables | ✅ | ✅ | ✅ | ✅ |
ggpiestats, ggbarstats |
Association between categorical variables | ✅ | ✅ | ❌ | ✅ |
ggpiestats, ggbarstats |
Equal proportions for categorical variable levels | ✅ | ✅ | ❌ | ✅ |
ggcoefstats |
Regression model coefficients | ✅ | ✅ | ✅ | ✅ |
ggcoefstats |
Random-effects meta-analysis | ✅ | ❌ | ✅ | ✅ |
Summary of Bayesian analysis
| Analysis | Hypothesis testing | Estimation |
|---|---|---|
| (one/two-sample) t-test | ✅ | ✅ |
| one-way ANOVA | ✅ | ✅ |
| correlation | ✅ | ✅ |
| (one/two-way) contingency table | ✅ | ✅ |
| random-effects meta-analysis | ✅ | ✅ |
For all statistical tests reported in the plots, the default template abides by the gold standard for statistical reporting. For example, here are results from Yuen’s test for trimmed means (robust t-test):

Statistical analysis is carried out by
{statsExpressions} package, and thus a summary table of all
the statistical tests currently supported across various functions can
be found in article for that package: https://indrajeetpatil.github.io/statsExpressions/articles/stats_details.html
ggbetweenstatsThis function creates either a violin plot, a box plot, or a mix of two for between-group or between-condition comparisons with results from statistical tests in the subtitle. The simplest function call looks like this-
set.seed(123)
ggbetweenstats(
data = iris,
x = Species,
y = Sepal.Length,
title = "Distribution of sepal length across Iris species"
)
Defaults return
✅ raw data + distributions
✅ descriptive statistics
✅
inferential statistics
✅ effect size + CIs
✅ pairwise
comparisons
✅ Bayesian hypothesis-testing
✅ Bayesian
estimation
A number of other arguments can be specified to make this plot even
more informative or change some of the default options. Additionally,
there is also a grouped_ variant of this function that
makes it easy to repeat the same operation across a
single grouping variable:
set.seed(123)
grouped_ggbetweenstats(
data = dplyr::filter(movies_long, genre %in% c("Action", "Comedy")),
x = mpaa,
y = length,
grouping.var = genre,
outlier.tagging = TRUE,
outlier.label = title,
outlier.coef = 2,
ggsignif.args = list(textsize = 4, tip_length = 0.01),
p.adjust.method = "bonferroni",
palette = "default_jama",
package = "ggsci",
plotgrid.args = list(nrow = 1),
annotation.args = list(title = "Differences in movie length by mpaa ratings for different genres")
)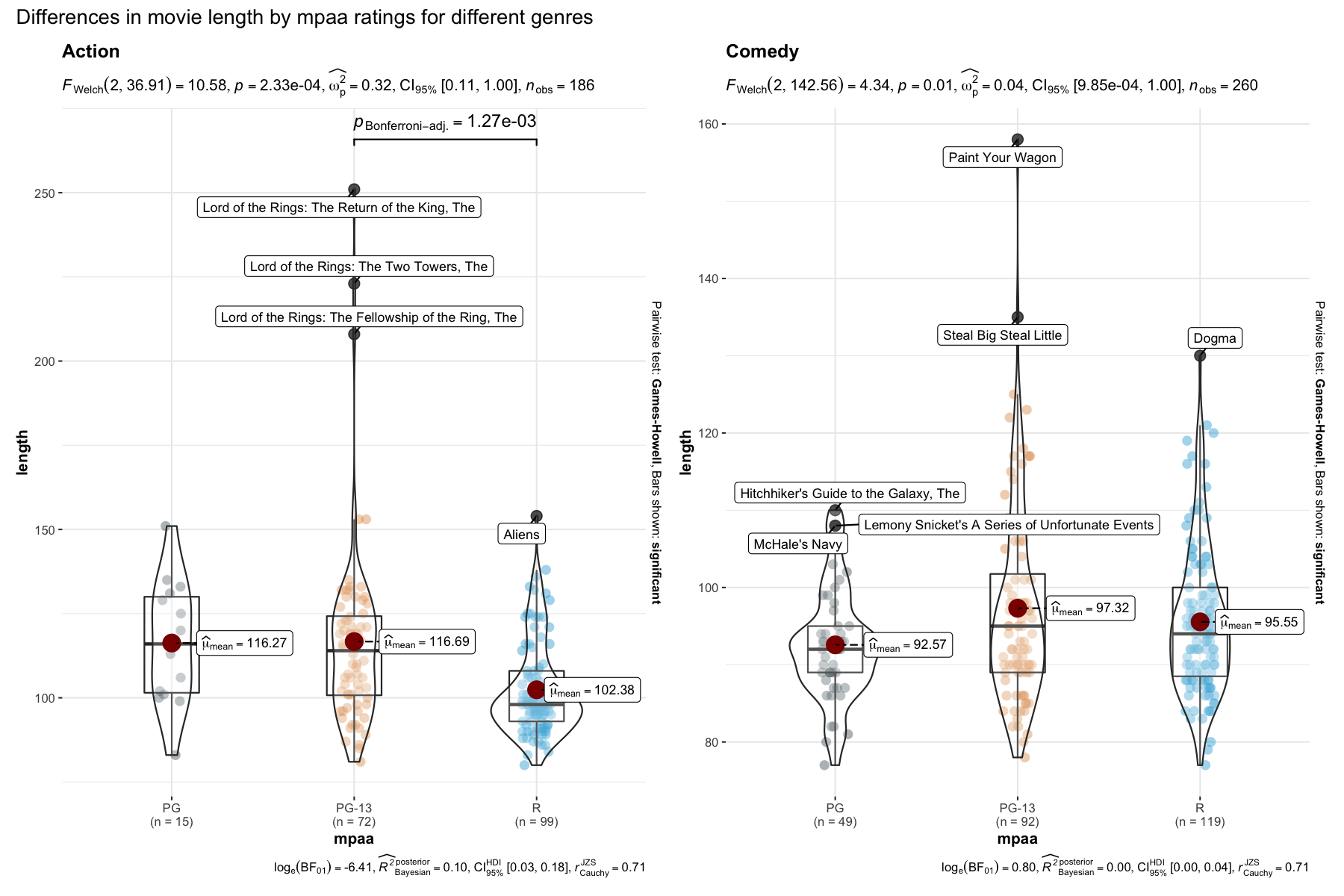
Note here that the function can be used to tag outliers!
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| raw data | ggplot2::geom_point |
point.args |
| box plot | ggplot2::geom_boxplot |
❌ |
| density plot | ggplot2::geom_violin |
violin.args |
| centrality measure point | ggplot2::geom_point |
centrality.point.args |
| centrality measure label | ggrepel::geom_label_repel |
centrality.label.args |
| outlier point | ggplot2::stat_boxplot |
❌ |
| outlier label | ggrepel::geom_label_repel |
outlier.label.args |
| pairwise comparisons | ggsignif::geom_signif |
ggsignif.args |
Central tendency measure
| Type | Measure | Function used |
|---|---|---|
| Parametric | mean | datawizard::describe_distribution |
| Non-parametric | median | datawizard::describe_distribution |
| Robust | trimmed mean | datawizard::describe_distribution |
| Bayesian | MAP (maximum a posteriori probability) estimate | datawizard::describe_distribution |
Hypothesis testing
| Type | No. of groups | Test | Function used |
|---|---|---|---|
| Parametric | > 2 | Fisher’s or Welch’s one-way ANOVA | stats::oneway.test |
| Non-parametric | > 2 | Kruskal–Wallis one-way ANOVA | stats::kruskal.test |
| Robust | > 2 | Heteroscedastic one-way ANOVA for trimmed means | WRS2::t1way |
| Bayes Factor | > 2 | Fisher’s ANOVA | BayesFactor::anovaBF |
| Parametric | 2 | Student’s or Welch’s t-test | stats::t.test |
| Non-parametric | 2 | Mann–Whitney U test | stats::wilcox.test |
| Robust | 2 | Yuen’s test for trimmed means | WRS2::yuen |
| Bayesian | 2 | Student’s t-test | BayesFactor::ttestBF |
Effect size estimation
| Type | No. of groups | Effect size | CI? | Function used |
|---|---|---|---|---|
| Parametric | > 2 | ✅ | effectsize::omega_squared,
effectsize::eta_squared |
|
| Non-parametric | > 2 | ✅ | effectsize::rank_epsilon_squared |
|
| Robust | > 2 | ✅ | WRS2::t1way |
|
| Bayes Factor | > 2 | ✅ | performance::r2_bayes |
|
| Parametric | 2 | Cohen’s d, Hedge’s g | ✅ | effectsize::cohens_d,
effectsize::hedges_g |
| Non-parametric | 2 | r (rank-biserial correlation) | ✅ | effectsize::rank_biserial |
| Robust | 2 | ✅ | WRS2::yuen.effect.ci |
|
| Bayesian | 2 | ✅ | bayestestR::describe_posterior |
Pairwise comparison tests
| Type | Equal variance? | Test | p-value adjustment? | Function used |
|---|---|---|---|---|
| Parametric | No | Games-Howell test | ✅ | PMCMRplus::gamesHowellTest |
| Parametric | Yes | Student’s t-test | ✅ | stats::pairwise.t.test |
| Non-parametric | No | Dunn test | ✅ | PMCMRplus::kwAllPairsDunnTest |
| Robust | No | Yuen’s trimmed means test | ✅ | WRS2::lincon |
| Bayesian | NA |
Student’s t-test | NA |
BayesFactor::ttestBF |
For more, see the ggbetweenstats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggbetweenstats.html
ggwithinstatsggbetweenstats function has an identical twin function
ggwithinstats for repeated measures designs that behaves in
the same fashion with a few minor tweaks introduced to properly
visualize the repeated measures design. As can be seen from an example
below, the only difference between the plot structure is that now the
group means are connected by paths to highlight the fact that these data
are paired with each other.
set.seed(123)
library(WRS2) ## for data
library(afex) ## to run anova
ggwithinstats(
data = WineTasting,
x = Wine,
y = Taste,
title = "Wine tasting"
)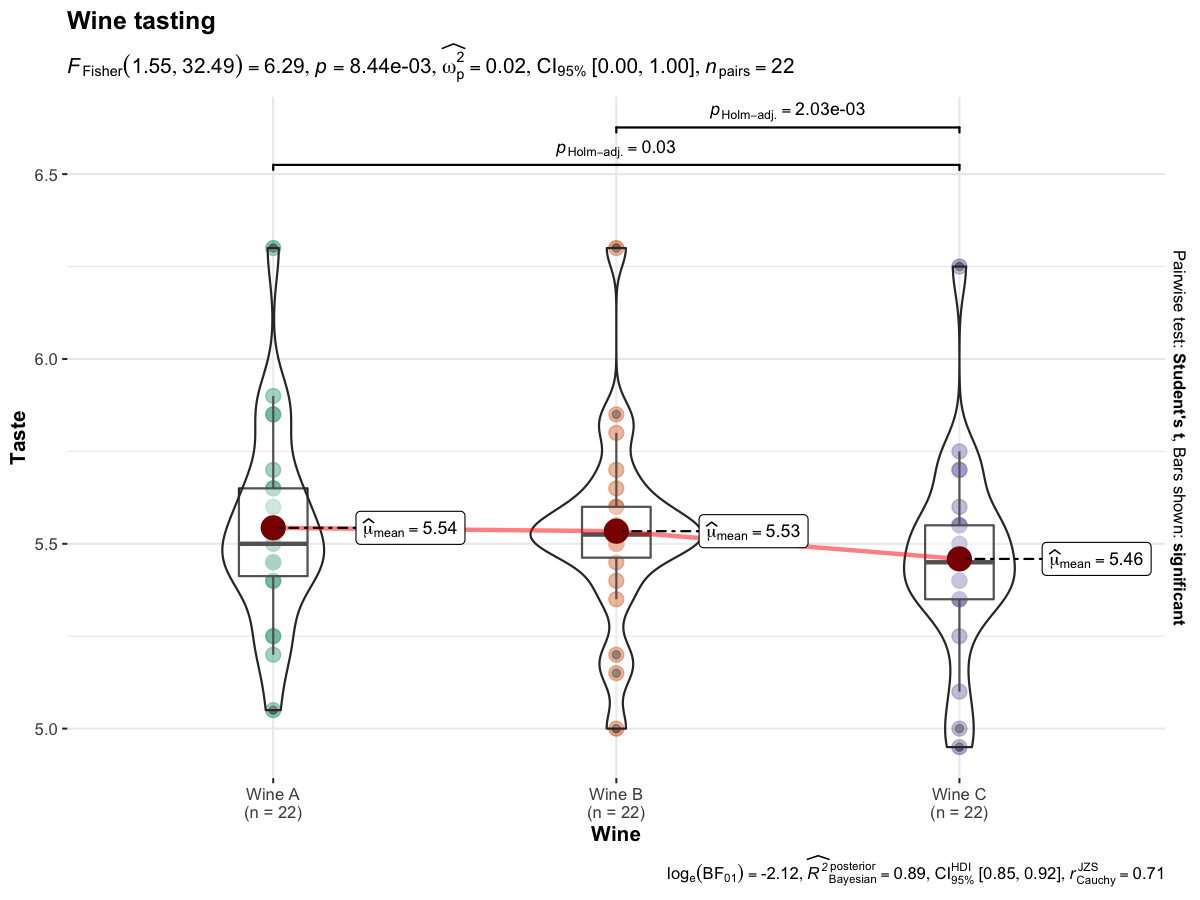
Defaults return
✅ raw data + distributions
✅ descriptive statistics
✅
inferential statistics
✅ effect size + CIs
✅ pairwise
comparisons
✅ Bayesian hypothesis-testing
✅ Bayesian
estimation
The central tendency measure displayed will depend on the statistics:
| Type | Measure | Function used |
|---|---|---|
| Parametric | mean | datawizard::describe_distribution |
| Non-parametric | median | datawizard::describe_distribution |
| Robust | trimmed mean | datawizard::describe_distribution |
| Bayesian | MAP estimate | datawizard::describe_distribution |
As with the ggbetweenstats, this function also has a
grouped_ variant that makes repeating the same analysis
across a single grouping variable quicker. We will see an example with
only repeated measurements-
set.seed(123)
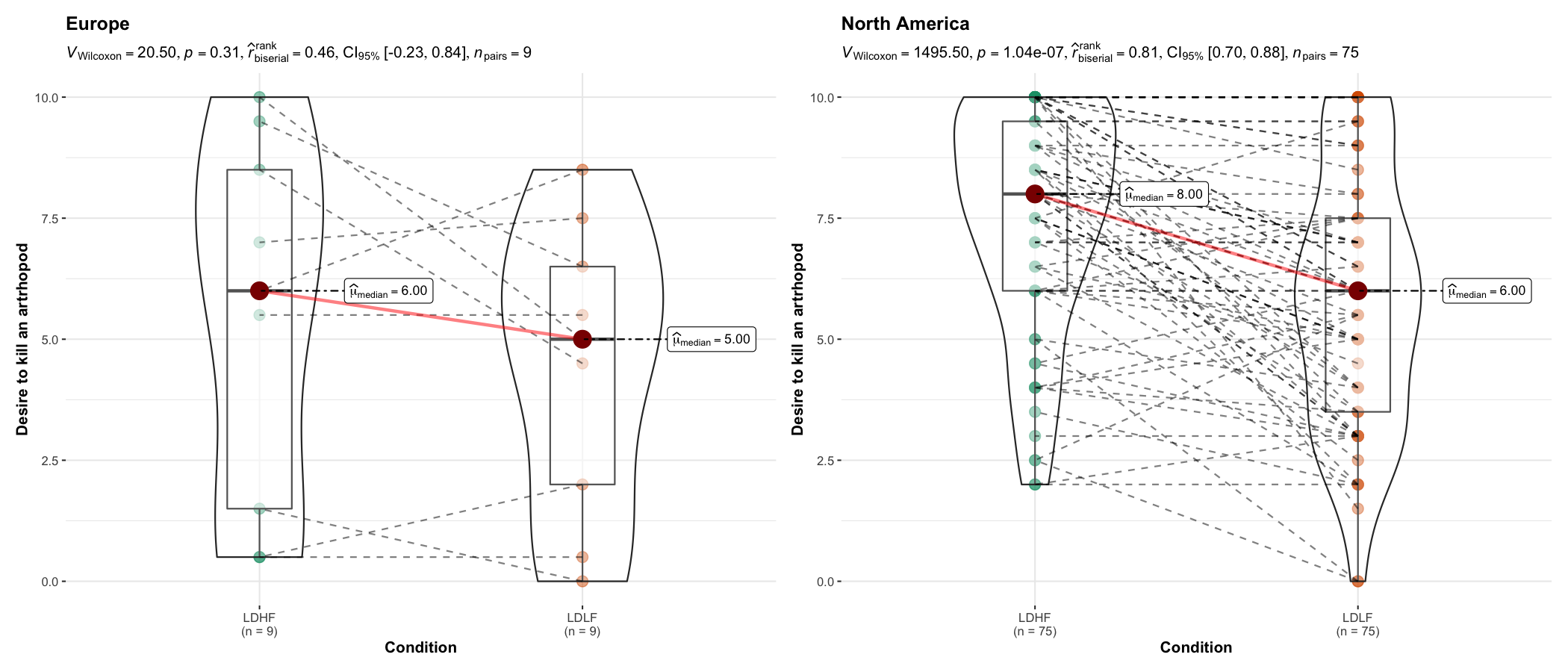
grouped_ggwithinstats(
data = dplyr::filter(bugs_long, region %in% c("Europe", "North America"), condition %in% c("LDLF", "LDHF")),
x = condition,
y = desire,
type = "np",
xlab = "Condition",
ylab = "Desire to kill an artrhopod",
grouping.var = region,
outlier.tagging = TRUE,
outlier.label = education
)
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| raw data | ggplot2::geom_point |
point.args |
| point path | ggplot2::geom_path |
point.path.args |
| box plot | ggplot2::geom_boxplot |
boxplot.args |
| density plot | ggplot2::geom_violin |
violin.args |
| centrality measure point | ggplot2::geom_point |
centrality.point.args |
| centrality measure point path | ggplot2::geom_path |
centrality.path.args |
| centrality measure label | ggrepel::geom_label_repel |
centrality.label.args |
| outlier point | ggplot2::stat_boxplot |
❌ |
| outlier label | ggrepel::geom_label_repel |
outlier.label.args |
| pairwise comparisons | ggsignif::geom_signif |
ggsignif.args |
Central tendency measure
| Type | Measure | Function used |
|---|---|---|
| Parametric | mean | datawizard::describe_distribution |
| Non-parametric | median | datawizard::describe_distribution |
| Robust | trimmed mean | datawizard::describe_distribution |
| Bayesian | MAP (maximum a posteriori probability) estimate | datawizard::describe_distribution |
Hypothesis testing
| Type | No. of groups | Test | Function used |
|---|---|---|---|
| Parametric | > 2 | One-way repeated measures ANOVA | afex::aov_ez |
| Non-parametric | > 2 | Friedman rank sum test | stats::friedman.test |
| Robust | > 2 | Heteroscedastic one-way repeated measures ANOVA for trimmed means | WRS2::rmanova |
| Bayes Factor | > 2 | One-way repeated measures ANOVA | BayesFactor::anovaBF |
| Parametric | 2 | Student’s t-test | stats::t.test |
| Non-parametric | 2 | Wilcoxon signed-rank test | stats::wilcox.test |
| Robust | 2 | Yuen’s test on trimmed means for dependent samples | WRS2::yuend |
| Bayesian | 2 | Student’s t-test | BayesFactor::ttestBF |
Effect size estimation
| Type | No. of groups | Effect size | CI? | Function used |
|---|---|---|---|---|
| Parametric | > 2 | ✅ | effectsize::omega_squared,
effectsize::eta_squared |
|
| Non-parametric | > 2 | ✅ | effectsize::kendalls_w |
|
| Robust | > 2 | ✅ | WRS2::wmcpAKP |
|
| Bayes Factor | > 2 | ✅ | performance::r2_bayes |
|
| Parametric | 2 | Cohen’s d, Hedge’s g | ✅ | effectsize::cohens_d,
effectsize::hedges_g |
| Non-parametric | 2 | r (rank-biserial correlation) | ✅ | effectsize::rank_biserial |
| Robust | 2 | ✅ | WRS2::wmcpAKP |
|
| Bayesian | 2 | ✅ | bayestestR::describe_posterior |
Pairwise comparison tests
| Type | Test | p-value adjustment? | Function used |
|---|---|---|---|
| Parametric | Student’s t-test | ✅ | stats::pairwise.t.test |
| Non-parametric | Durbin-Conover test | ✅ | PMCMRplus::durbinAllPairsTest |
| Robust | Yuen’s trimmed means test | ✅ | WRS2::rmmcp |
| Bayesian | Student’s t-test | ❌ | BayesFactor::ttestBF |
For more, see the ggwithinstats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggwithinstats.html
gghistostatsTo visualize the distribution of a single variable and check if its
mean is significantly different from a specified value with a one-sample
test, gghistostats can be used.
set.seed(123)
gghistostats(
data = ggplot2::msleep,
x = awake,
title = "Amount of time spent awake",
test.value = 12,
binwidth = 1
)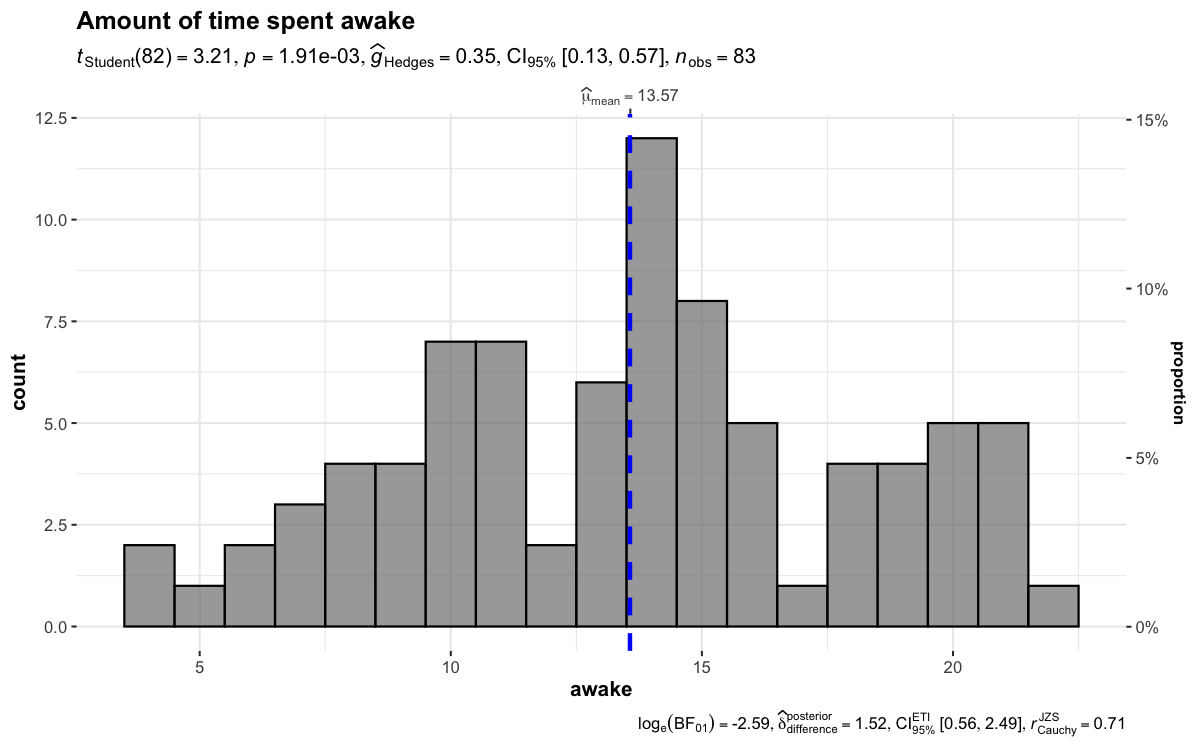
Defaults return
✅ counts + proportion for bins
✅ descriptive statistics
✅
inferential statistics
✅ effect size + CIs
✅ Bayesian
hypothesis-testing
✅ Bayesian estimation
There is also a grouped_ variant of this function that
makes it easy to repeat the same operation across a
single grouping variable:
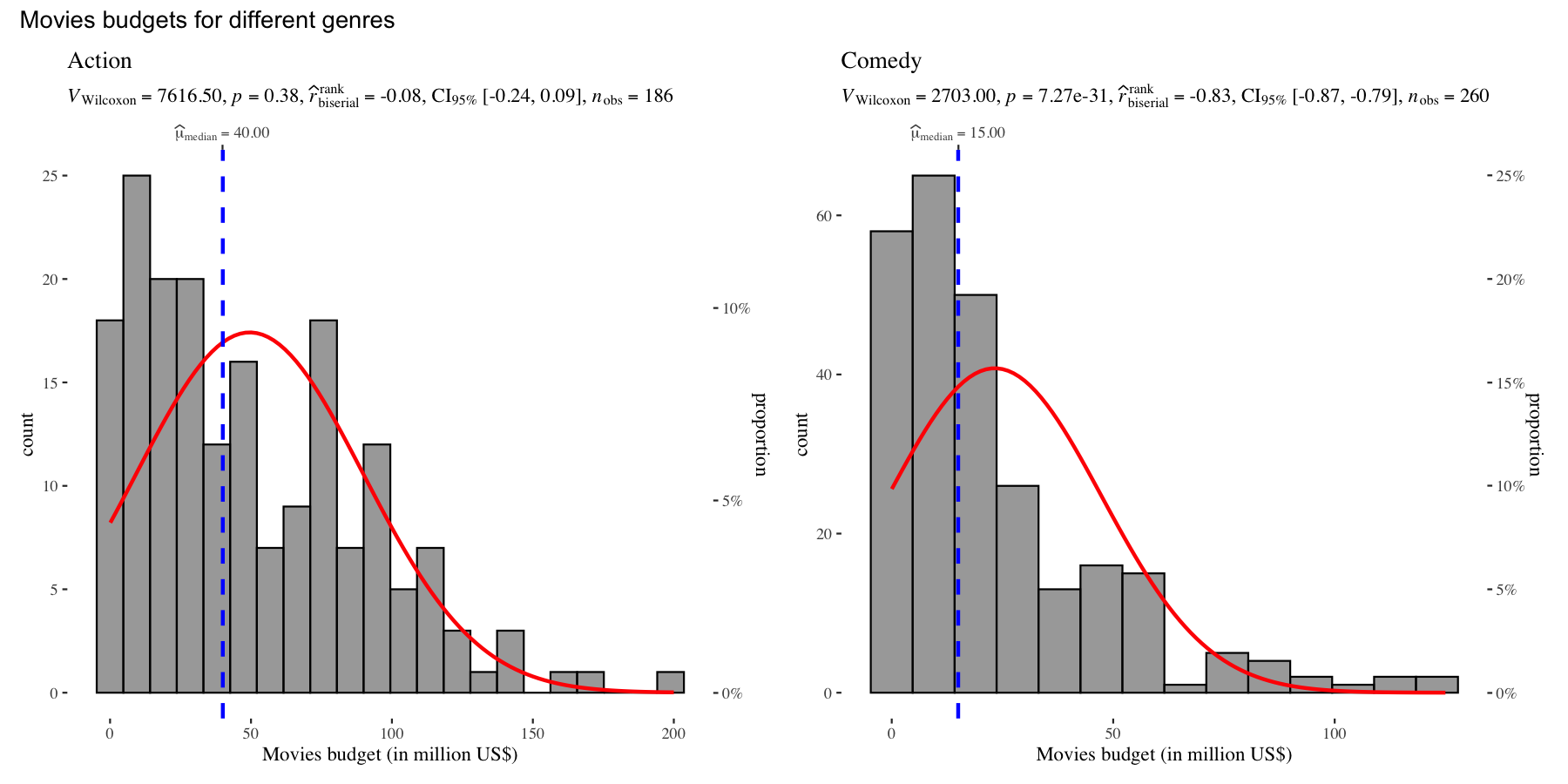
set.seed(123)
grouped_gghistostats(
data = dplyr::filter(movies_long, genre %in% c("Action", "Comedy")),
x = budget,
test.value = 50,
type = "nonparametric",
xlab = "Movies budget (in million US$)",
grouping.var = genre,
normal.curve = TRUE,
normal.curve.args = list(color = "red", size = 1),
ggtheme = ggthemes::theme_tufte(),
## modify the defaults from `{ggstatsplot}` for each plot
plotgrid.args = list(nrow = 1),
annotation.args = list(title = "Movies budgets for different genres")
)
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| histogram bin | ggplot2::stat_bin |
bin.args |
| centrality measure line | ggplot2::geom_vline |
centrality.line.args |
| normality curve | ggplot2::stat_function |
normal.curve.args |
Central tendency measure
| Type | Measure | Function used |
|---|---|---|
| Parametric | mean | datawizard::describe_distribution |
| Non-parametric | median | datawizard::describe_distribution |
| Robust | trimmed mean | datawizard::describe_distribution |
| Bayesian | MAP (maximum a posteriori probability) estimate | datawizard::describe_distribution |
Hypothesis testing
| Type | Test | Function used |
|---|---|---|
| Parametric | One-sample Student’s t-test | stats::t.test |
| Non-parametric | One-sample Wilcoxon test | stats::wilcox.test |
| Robust | Bootstrap-t method for one-sample test | WRS2::trimcibt |
| Bayesian | One-sample Student’s t-test | BayesFactor::ttestBF |
Effect size estimation
| Type | Effect size | CI? | Function used |
|---|---|---|---|
| Parametric | Cohen’s d, Hedge’s g | ✅ | effectsize::cohens_d,
effectsize::hedges_g |
| Non-parametric | r (rank-biserial correlation) | ✅ | effectsize::rank_biserial |
| Robust | trimmed mean | ✅ | WRS2::trimcibt |
| Bayes Factor | ✅ | bayestestR::describe_posterior |
For more, including information about the variant of this function
grouped_gghistostats, see the gghistostats
vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/gghistostats.html
ggdotplotstatsThis function is similar to gghistostats, but is
intended to be used when the numeric variable also has a label.
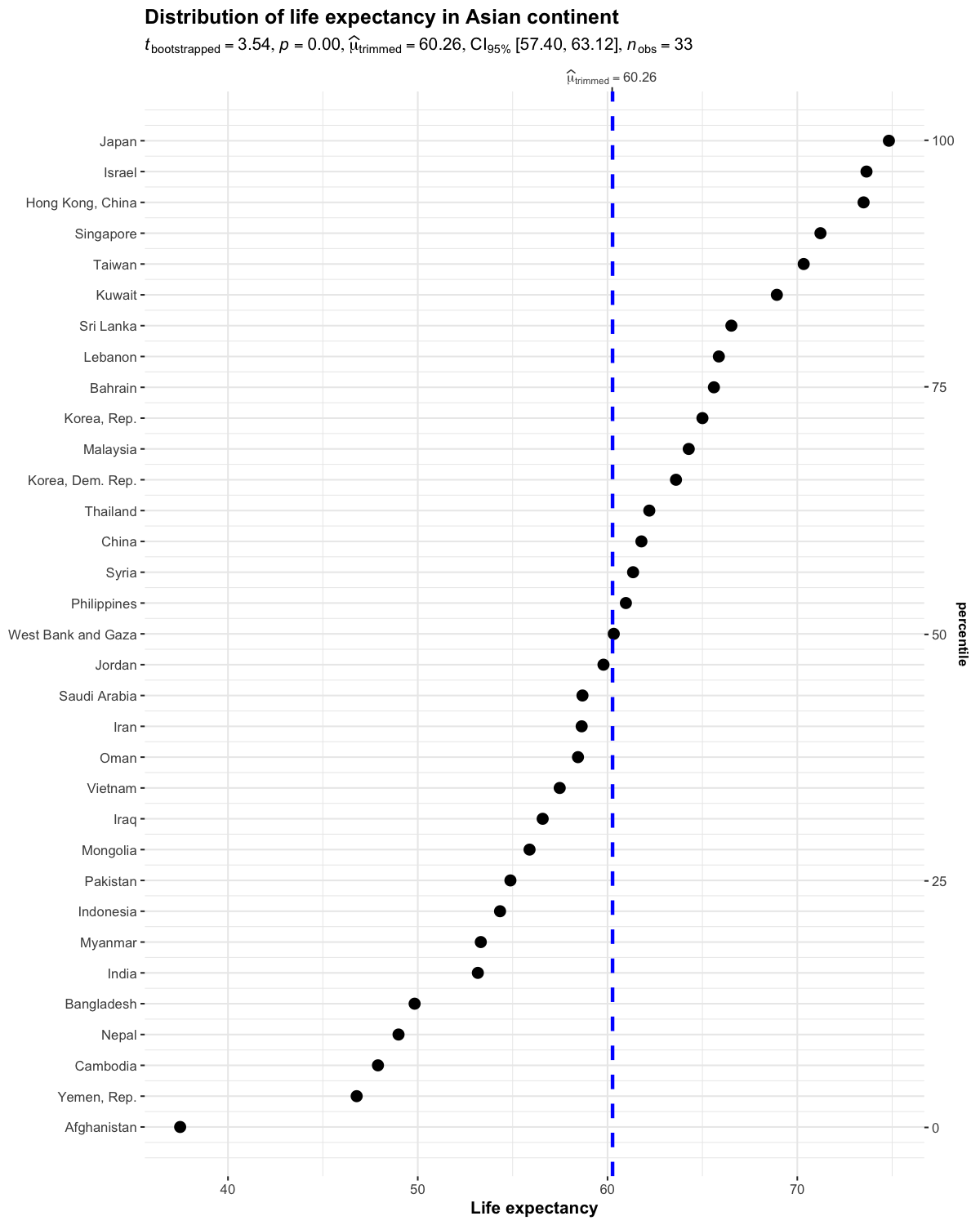
set.seed(123)
ggdotplotstats(
data = dplyr::filter(gapminder::gapminder, continent == "Asia"),
y = country,
x = lifeExp,
test.value = 55,
type = "robust",
title = "Distribution of life expectancy in Asian continent",
xlab = "Life expectancy"
)
Defaults return
✅ descriptives (mean + sample size)
✅ inferential statistics
✅ effect size + CIs
✅ Bayesian hypothesis-testing
✅
Bayesian estimation
As with the rest of the functions in this package, there is also a
grouped_ variant of this function to facilitate looping the
same operation for all levels of a single grouping variable.
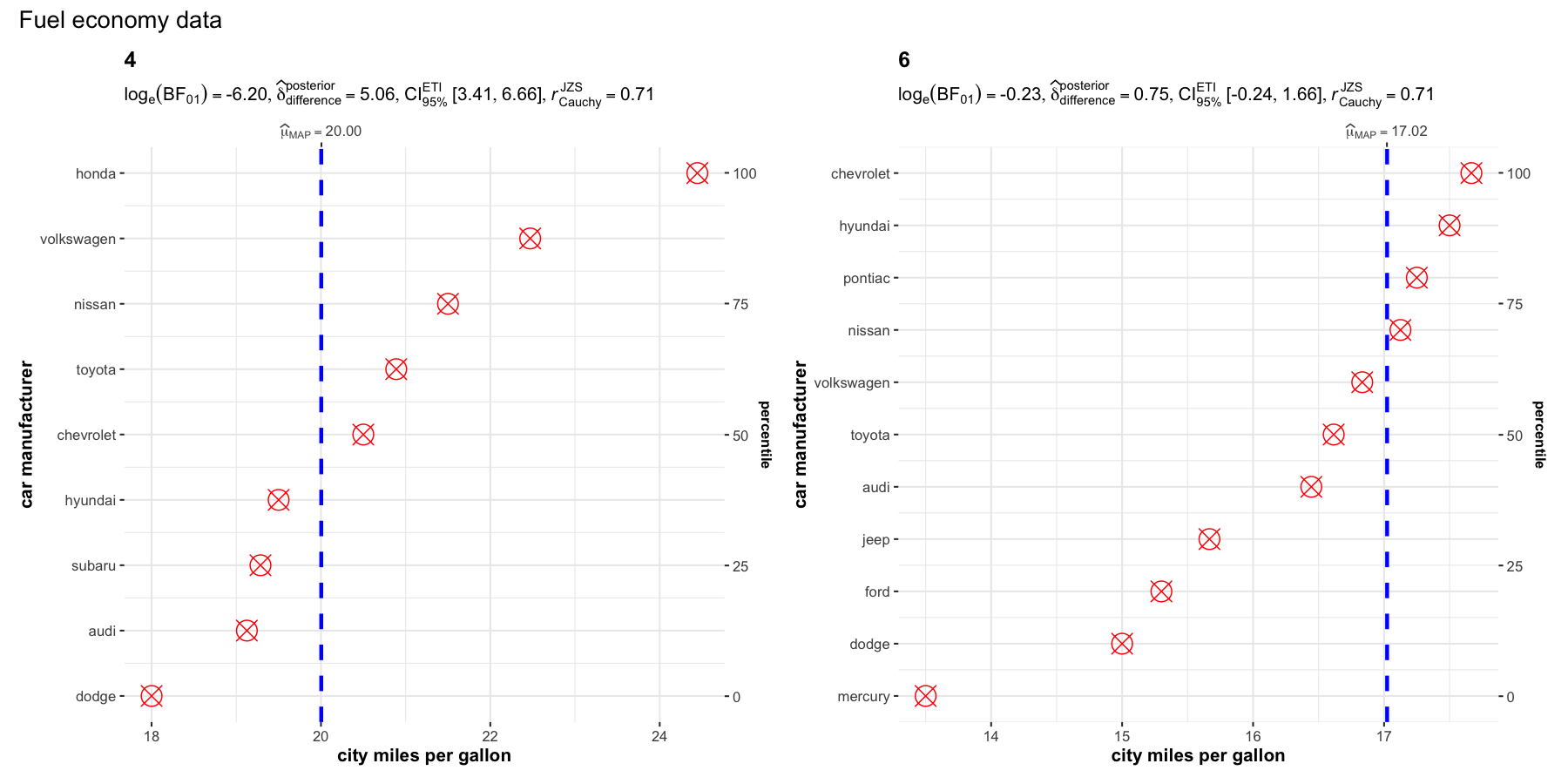
set.seed(123)
grouped_ggdotplotstats(
data = dplyr::filter(ggplot2::mpg, cyl %in% c("4", "6")),
x = cty,
y = manufacturer,
type = "bayes",
xlab = "city miles per gallon",
ylab = "car manufacturer",
grouping.var = cyl,
test.value = 15.5,
point.args = list(color = "red", size = 5, shape = 13),
annotation.args = list(title = "Fuel economy data")
)
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| raw data | ggplot2::geom_point |
point.args |
| centrality measure line | ggplot2::geom_vline |
centrality.line.args |
Central tendency measure
| Type | Measure | Function used |
|---|---|---|
| Parametric | mean | datawizard::describe_distribution |
| Non-parametric | median | datawizard::describe_distribution |
| Robust | trimmed mean | datawizard::describe_distribution |
| Bayesian | MAP (maximum a posteriori probability) estimate | datawizard::describe_distribution |
Hypothesis testing
| Type | Test | Function used |
|---|---|---|
| Parametric | One-sample Student’s t-test | stats::t.test |
| Non-parametric | One-sample Wilcoxon test | stats::wilcox.test |
| Robust | Bootstrap-t method for one-sample test | WRS2::trimcibt |
| Bayesian | One-sample Student’s t-test | BayesFactor::ttestBF |
Effect size estimation
| Type | Effect size | CI? | Function used |
|---|---|---|---|
| Parametric | Cohen’s d, Hedge’s g | ✅ | effectsize::cohens_d,
effectsize::hedges_g |
| Non-parametric | r (rank-biserial correlation) | ✅ | effectsize::rank_biserial |
| Robust | trimmed mean | ✅ | WRS2::trimcibt |
| Bayes Factor | ✅ | bayestestR::describe_posterior |
ggscatterstatsThis function creates a scatterplot with marginal distributions overlaid on the axes and results from statistical tests in the subtitle:
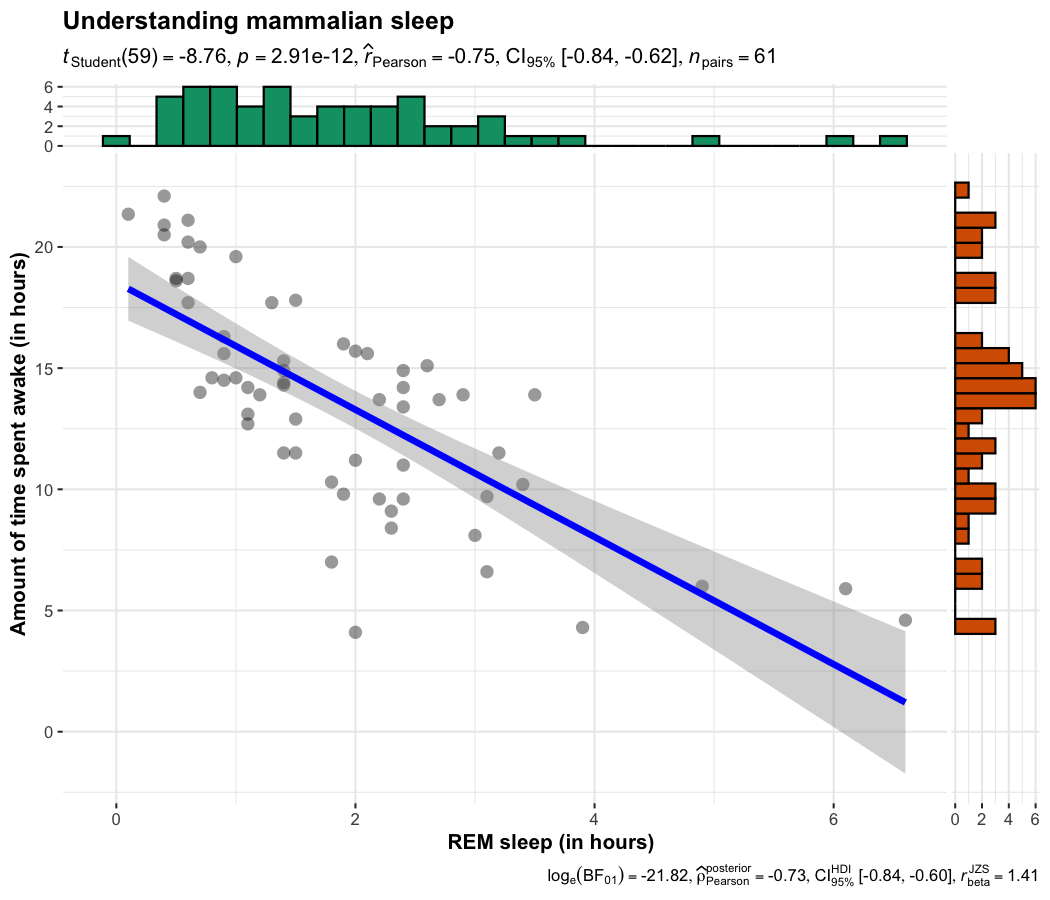
ggscatterstats(
data = ggplot2::msleep,
x = sleep_rem,
y = awake,
xlab = "REM sleep (in hours)",
ylab = "Amount of time spent awake (in hours)",
title = "Understanding mammalian sleep"
)
Defaults return
✅ raw data + distributions
✅ marginal distributions
✅
inferential statistics
✅ effect size + CIs
✅ Bayesian
hypothesis-testing
✅ Bayesian estimation
There is also a grouped_ variant of this function that
makes it easy to repeat the same operation across a
single grouping variable.
set.seed(123)
grouped_ggscatterstats(
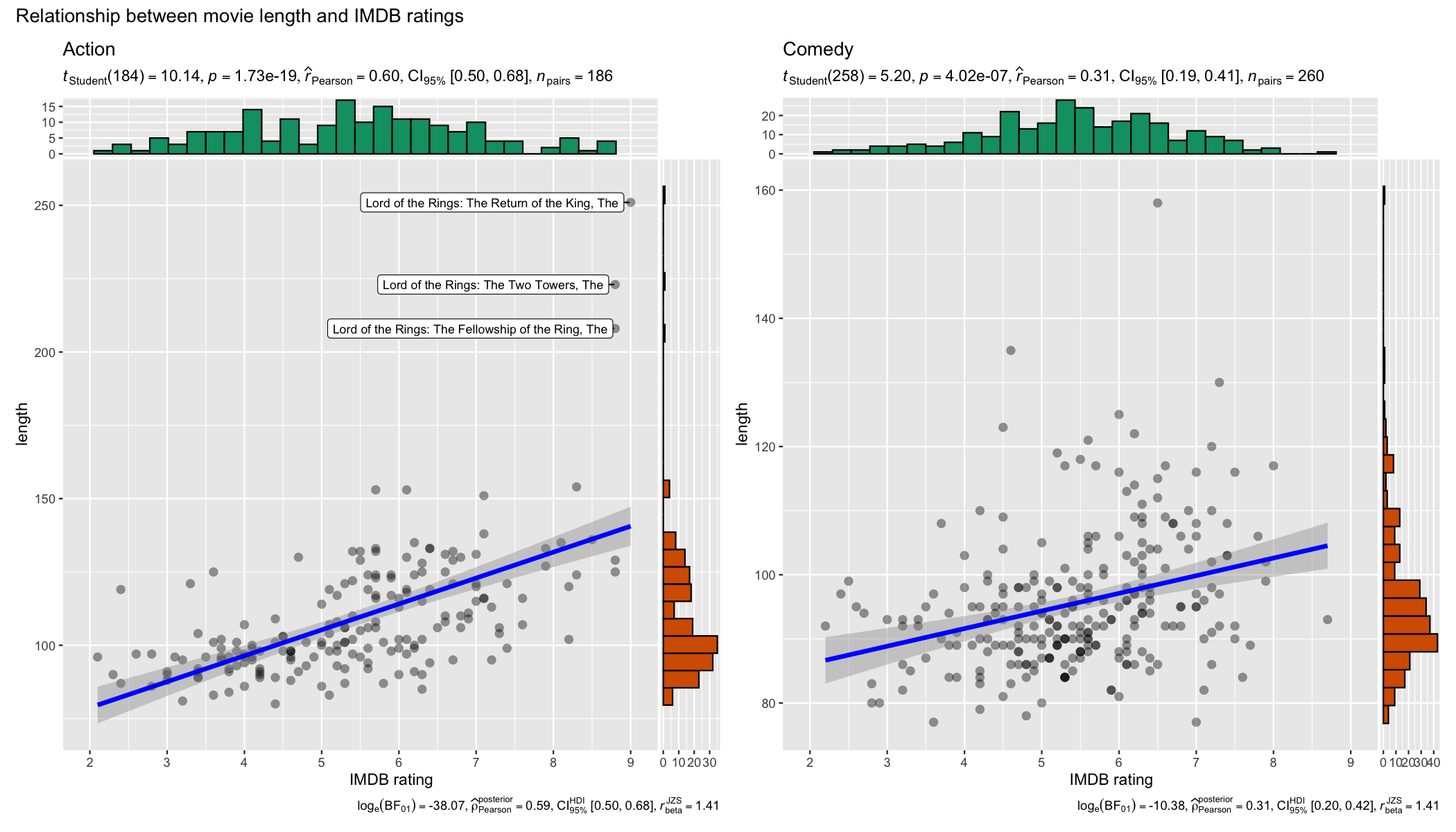
data = dplyr::filter(movies_long, genre %in% c("Action", "Comedy")),
x = rating,
y = length,
grouping.var = genre,
label.var = title,
label.expression = length > 200,
xlab = "IMDB rating",
ggtheme = ggplot2::theme_grey(),
ggplot.component = list(ggplot2::scale_x_continuous(breaks = seq(2, 9, 1), limits = (c(2, 9)))),
plotgrid.args = list(nrow = 1),
annotation.args = list(title = "Relationship between movie length and IMDB ratings")
)
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| raw data | ggplot2::geom_point |
point.args |
| labels for raw data | ggrepel::geom_label_repel |
point.label.args |
| smooth line | ggplot2::geom_smooth |
smooth.line.args |
| marginal histograms | ggside::geom_xsidehistogram,
ggside::geom_ysidehistogram |
xsidehistogram.args,
ysidehistogram.args |
Hypothesis testing and Effect size estimation
| Type | Test | CI? | Function used |
|---|---|---|---|
| Parametric | Pearson’s correlation coefficient | ✅ | correlation::correlation |
| Non-parametric | Spearman’s rank correlation coefficient | ✅ | correlation::correlation |
| Robust | Winsorized Pearson correlation coefficient | ✅ | correlation::correlation |
| Bayesian | Pearson’s correlation coefficient | ✅ | correlation::correlation |
For more, see the ggscatterstats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggscatterstats.html
ggcorrmatggcorrmat makes a correlalogram (a matrix of correlation
coefficients) with minimal amount of code. Just sticking to the defaults
itself produces publication-ready correlation matrices. But, for the
sake of exploring the available options, let’s change some of the
defaults. For example, multiple aesthetics-related arguments can be
modified to change the appearance of the correlation matrix.
set.seed(123)
## as a default this function outputs a correlation matrix plot
ggcorrmat(
data = ggplot2::msleep,
colors = c("#B2182B", "white", "#4D4D4D"),
title = "Correlalogram for mammals sleep dataset",
subtitle = "sleep units: hours; weight units: kilograms"
)
Defaults return
✅ effect size + significance
✅ careful handling of
NAs
If there are NAs present in the selected variables, the
legend will display minimum, median, and maximum number of pairs used
for correlation tests.
There is also a grouped_ variant of this function that
makes it easy to repeat the same operation across a
single grouping variable:
set.seed(123)
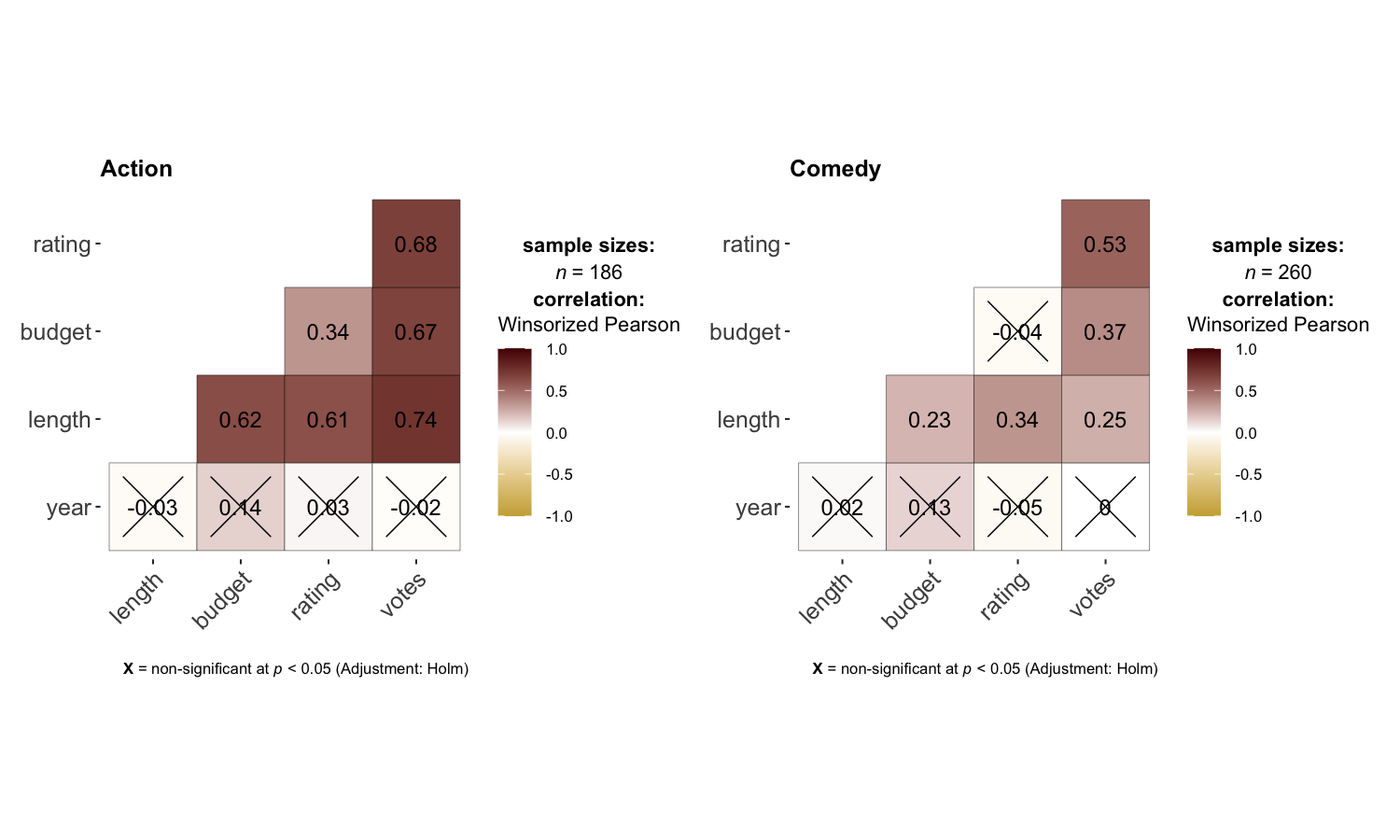
grouped_ggcorrmat(
data = dplyr::filter(movies_long, genre %in% c("Action", "Comedy")),
type = "robust",
colors = c("#cbac43", "white", "#550000"),
grouping.var = genre,
matrix.type = "lower"
)
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| correlation matrix | ggcorrplot::ggcorrplot |
ggcorrplot.args |
Hypothesis testing and Effect size estimation
| Type | Test | CI? | Function used |
|---|---|---|---|
| Parametric | Pearson’s correlation coefficient | ✅ | correlation::correlation |
| Non-parametric | Spearman’s rank correlation coefficient | ✅ | correlation::correlation |
| Robust | Winsorized Pearson correlation coefficient | ✅ | correlation::correlation |
| Bayesian | Pearson’s correlation coefficient | ✅ | correlation::correlation |
For examples and more information, see the ggcorrmat
vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggcorrmat.html
ggpiestatsThis function creates a pie chart for categorical or nominal variables with results from contingency table analysis (Pearson’s chi-squared test for between-subjects design and McNemar’s chi-squared test for within-subjects design) included in the subtitle of the plot. If only one categorical variable is entered, results from one-sample proportion test (i.e., a chi-squared goodness of fit test) will be displayed as a subtitle.
To study an interaction between two categorical variables:
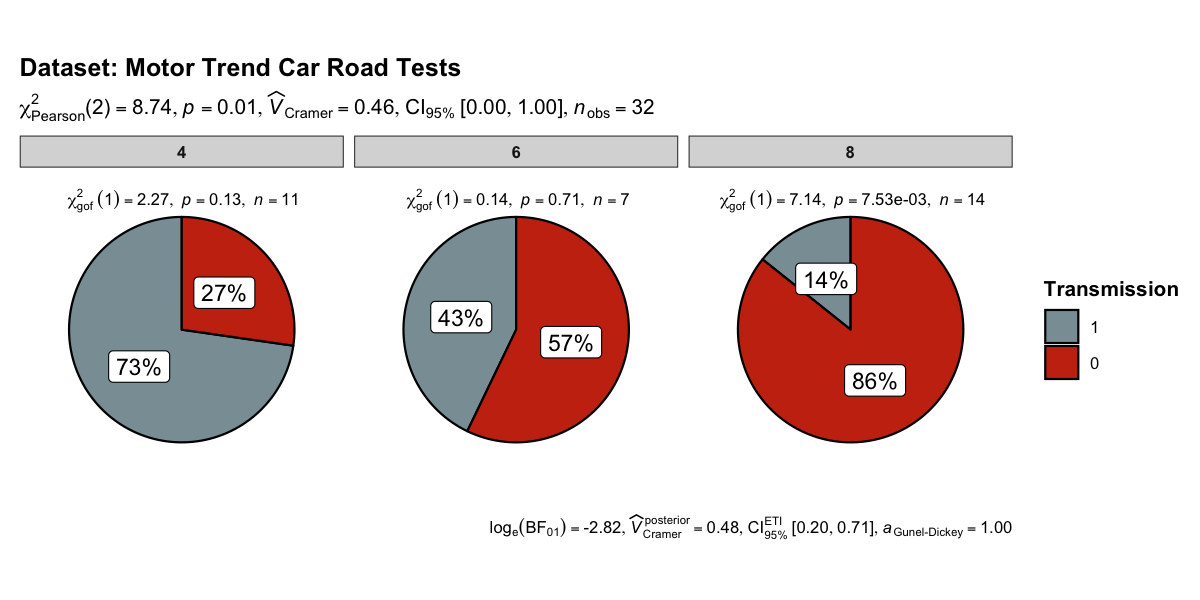
set.seed(123)
ggpiestats(
data = mtcars,
x = am,
y = cyl,
package = "wesanderson",
palette = "Royal1",
title = "Dataset: Motor Trend Car Road Tests",
legend.title = "Transmission"
)
Defaults return
✅ descriptives (frequency + %s)
✅ inferential statistics
✅ effect size + CIs
✅ Goodness-of-fit tests
✅ Bayesian
hypothesis-testing
✅ Bayesian estimation
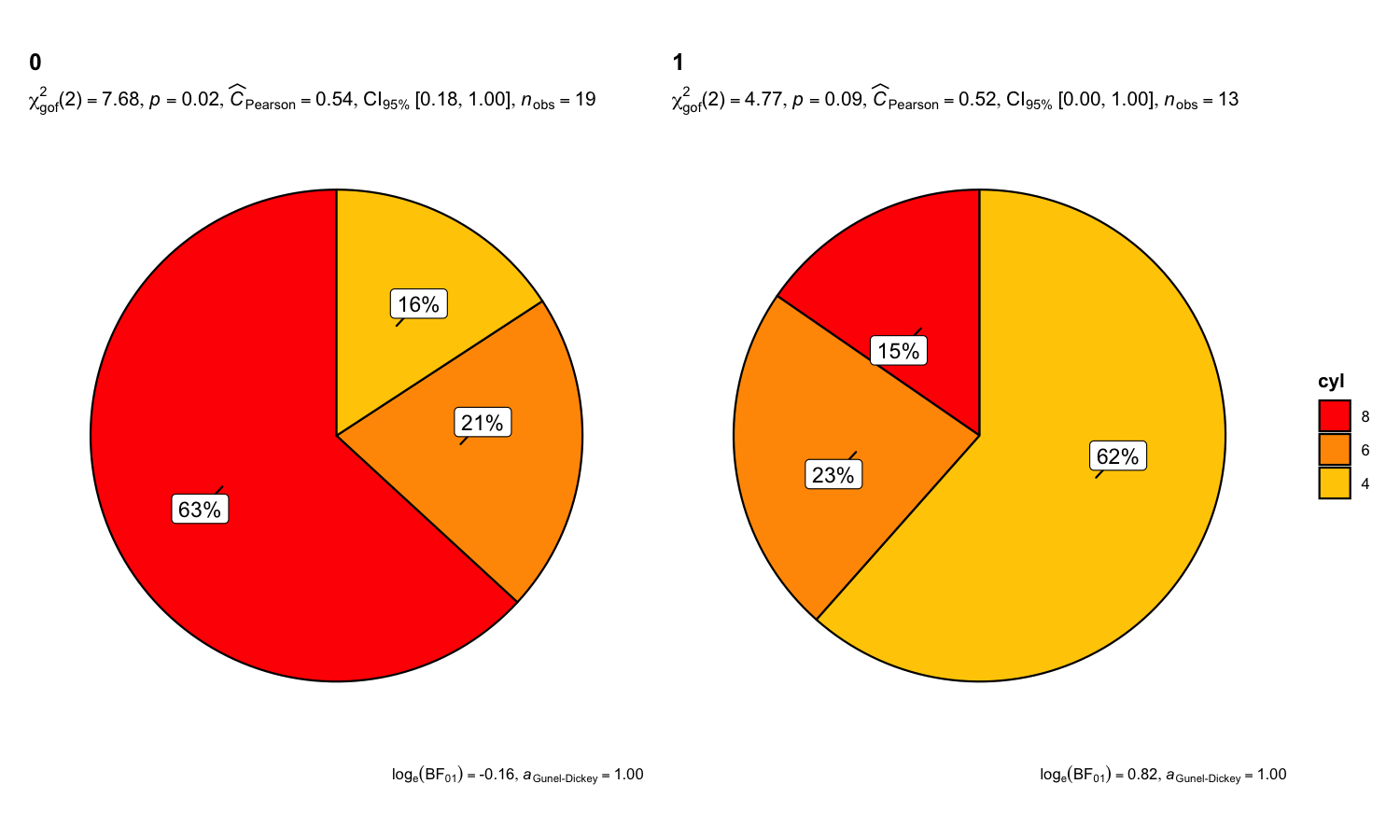
There is also a grouped_ variant of this function that
makes it easy to repeat the same operation across a
single grouping variable. Following example is a case
where the theoretical question is about proportions for different levels
of a single nominal variable:
set.seed(123)
grouped_ggpiestats(
data = mtcars,
x = cyl,
grouping.var = am,
label.repel = TRUE,
package = "ggsci",
palette = "default_ucscgb"
)
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| pie slices | ggplot2::geom_col |
❌ |
| descriptive labels | ggplot2::geom_label/ggrepel::geom_label_repel |
label.args |
two-way table
Hypothesis testing
| Type | Design | Test | Function used |
|---|---|---|---|
| Parametric/Non-parametric | Unpaired | Pearson’s |
stats::chisq.test |
| Bayesian | Unpaired | Bayesian Pearson’s |
BayesFactor::contingencyTableBF |
| Parametric/Non-parametric | Paired | McNemar’s |
stats::mcnemar.test |
| Bayesian | Paired | ❌ | ❌ |
Effect size estimation
| Type | Design | Effect size | CI? | Function used |
|---|---|---|---|---|
| Parametric/Non-parametric | Unpaired | Cramer’s |
✅ | effectsize::cramers_v |
| Bayesian | Unpaired | Cramer’s |
✅ | effectsize::cramers_v |
| Parametric/Non-parametric | Paired | Cohen’s |
✅ | effectsize::cohens_g |
| Bayesian | Paired | ❌ | ❌ | ❌ |
one-way table
Hypothesis testing
| Type | Test | Function used |
|---|---|---|
| Parametric/Non-parametric | Goodness of fit |
stats::chisq.test |
| Bayesian | Bayesian Goodness of fit |
(custom) |
Effect size estimation
| Type | Effect size | CI? | Function used |
|---|---|---|---|
| Parametric/Non-parametric | Pearson’s |
✅ | effectsize::pearsons_c |
| Bayesian | ❌ | ❌ | ❌ |
For more, see the ggpiestats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggpiestats.html
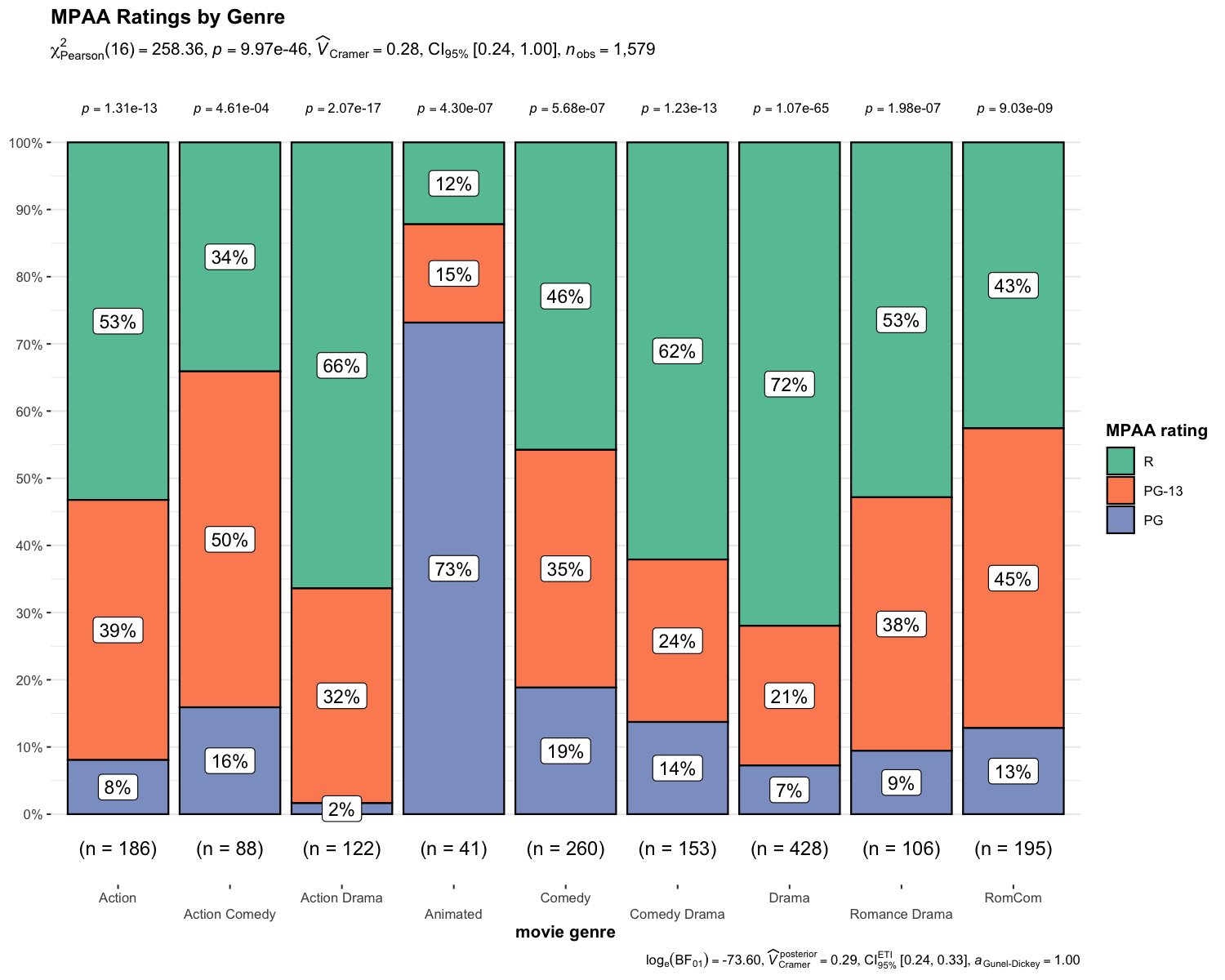
ggbarstatsIn case you are not a fan of pie charts (for very good reasons), you
can alternatively use ggbarstats function which has a
similar syntax.
N.B. The p-values from one-sample proportion test are displayed on top of each bar.
set.seed(123)
library(ggplot2)
ggbarstats(
data = movies_long,
x = mpaa,
y = genre,
title = "MPAA Ratings by Genre",
xlab = "movie genre",
legend.title = "MPAA rating",
ggplot.component = list(ggplot2::scale_x_discrete(guide = ggplot2::guide_axis(n.dodge = 2))),
palette = "Set2"
)
Defaults return
✅ descriptives (frequency + %s)
✅ inferential statistics
✅ effect size + CIs
✅ Goodness-of-fit tests
✅ Bayesian
hypothesis-testing
✅ Bayesian estimation
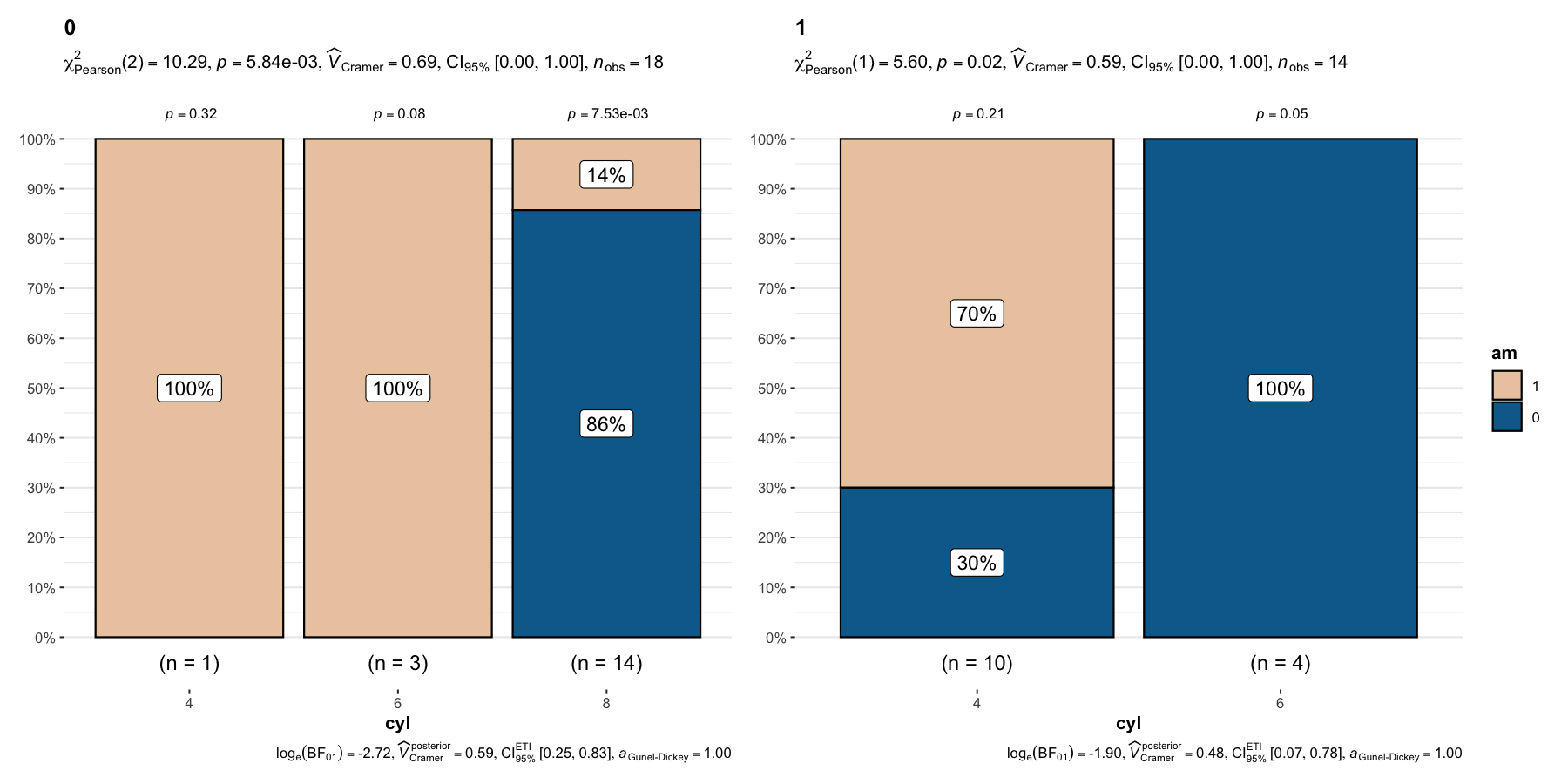
And, needless to say, there is also a grouped_ variant
of this function-
## setup
set.seed(123)
grouped_ggbarstats(
data = mtcars,
x = am,
y = cyl,
grouping.var = vs,
package = "wesanderson",
palette = "Darjeeling2" # ,
# ggtheme = ggthemes::theme_tufte(base_size = 12)
)
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| bars | ggplot2::geom_bar |
❌ |
| descriptive labels | ggplot2::geom_label |
label.args |
two-way table
Hypothesis testing
| Type | Design | Test | Function used |
|---|---|---|---|
| Parametric/Non-parametric | Unpaired | Pearson’s |
stats::chisq.test |
| Bayesian | Unpaired | Bayesian Pearson’s |
BayesFactor::contingencyTableBF |
| Parametric/Non-parametric | Paired | McNemar’s |
stats::mcnemar.test |
| Bayesian | Paired | ❌ | ❌ |
Effect size estimation
| Type | Design | Effect size | CI? | Function used |
|---|---|---|---|---|
| Parametric/Non-parametric | Unpaired | Cramer’s |
✅ | effectsize::cramers_v |
| Bayesian | Unpaired | Cramer’s |
✅ | effectsize::cramers_v |
| Parametric/Non-parametric | Paired | Cohen’s |
✅ | effectsize::cohens_g |
| Bayesian | Paired | ❌ | ❌ | ❌ |
one-way table
Hypothesis testing
| Type | Test | Function used |
|---|---|---|
| Parametric/Non-parametric | Goodness of fit |
stats::chisq.test |
| Bayesian | Bayesian Goodness of fit |
(custom) |
Effect size estimation
| Type | Effect size | CI? | Function used |
|---|---|---|---|
| Parametric/Non-parametric | Pearson’s |
✅ | effectsize::pearsons_c |
| Bayesian | ❌ | ❌ | ❌ |
ggcoefstatsThe function ggcoefstats generates
dot-and-whisker plots for regression models saved in a
tidy data frame. The tidy data frames are prepared using
parameters::model_parameters(). Additionally, if available,
the model summary indices are also extracted from
performance::model_performance().
Although the statistical models displayed in the plot may differ based on the class of models being investigated, there are few aspects of the plot that will be invariant across models:
The dot-whisker plot contains a dot representing the
estimate and their confidence
intervals (95% is the default). The estimate can
either be effect sizes (for tests that depend on the
F-statistic) or regression coefficients (for tests with
t-, -, and
z-statistic), etc. The
function will, by default, display a helpful x-axis label
that should clear up what estimates are being displayed. The confidence
intervals can sometimes be asymmetric if bootstrapping was
used.
The label attached to dot will provide more details from the statistical test carried out and it will typically contain estimate, statistic, and p-value.e
The caption will contain diagnostic information, if available, about models that can be useful for model selection: The smaller the Akaike’s Information Criterion (AIC) and the Bayesian Information Criterion (BIC) values, the “better” the model is.
The output of this function will be a {ggplot2}
object and, thus, it can be further modified (e.g. change themes) with
{ggplot2} functions.
set.seed(123)
## model
mod <- stats::lm(formula = mpg ~ am * cyl, data = mtcars)
ggcoefstats(mod)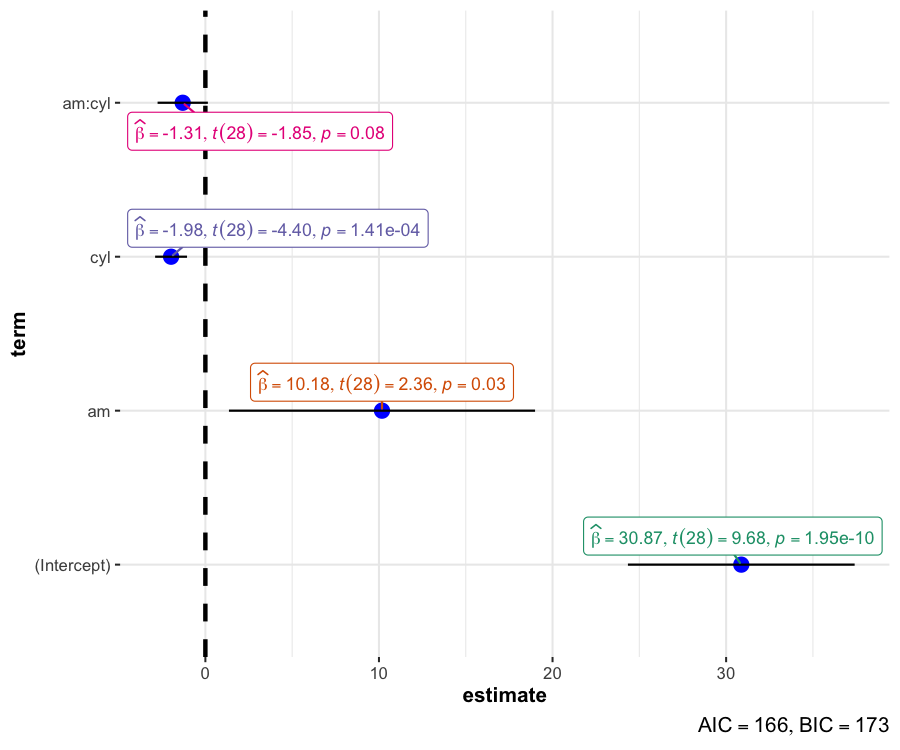
Defaults return
✅ inferential statistics
✅ estimate + CIs
✅ model
summary (AIC and BIC)
Most of the regression models that are supported in the underlying
packages are also supported by ggcoefstats.
insight::supported_models()
#> [1] "aareg" "afex_aov"
#> [3] "AKP" "Anova.mlm"
#> [5] "anova.rms" "aov"
#> [7] "aovlist" "Arima"
#> [9] "averaging" "bamlss"
#> [11] "bamlss.frame" "bayesQR"
#> [13] "bayesx" "BBmm"
#> [15] "BBreg" "bcplm"
#> [17] "betamfx" "betaor"
#> [19] "betareg" "BFBayesFactor"
#> [21] "bfsl" "BGGM"
#> [23] "bife" "bifeAPEs"
#> [25] "bigglm" "biglm"
#> [27] "blavaan" "blrm"
#> [29] "bracl" "brglm"
#> [31] "brmsfit" "brmultinom"
#> [33] "btergm" "censReg"
#> [35] "cgam" "cgamm"
#> [37] "cglm" "clm"
#> [39] "clm2" "clmm"
#> [41] "clmm2" "clogit"
#> [43] "coeftest" "complmrob"
#> [45] "confusionMatrix" "coxme"
#> [47] "coxph" "coxph.penal"
#> [49] "coxr" "cpglm"
#> [51] "cpglmm" "crch"
#> [53] "crq" "crqs"
#> [55] "crr" "dep.effect"
#> [57] "DirichletRegModel" "drc"
#> [59] "eglm" "elm"
#> [61] "epi.2by2" "ergm"
#> [63] "feglm" "feis"
#> [65] "felm" "fitdistr"
#> [67] "fixest" "flexsurvreg"
#> [69] "gam" "Gam"
#> [71] "gamlss" "gamm"
#> [73] "gamm4" "garch"
#> [75] "gbm" "gee"
#> [77] "geeglm" "glht"
#> [79] "glimML" "glm"
#> [81] "Glm" "glmm"
#> [83] "glmmadmb" "glmmPQL"
#> [85] "glmmTMB" "glmrob"
#> [87] "glmRob" "glmx"
#> [89] "gls" "gmnl"
#> [91] "HLfit" "htest"
#> [93] "hurdle" "iv_robust"
#> [95] "ivFixed" "ivprobit"
#> [97] "ivreg" "lavaan"
#> [99] "lm" "lm_robust"
#> [101] "lme" "lmerMod"
#> [103] "lmerModLmerTest" "lmodel2"
#> [105] "lmrob" "lmRob"
#> [107] "logistf" "logitmfx"
#> [109] "logitor" "LORgee"
#> [111] "lqm" "lqmm"
#> [113] "lrm" "manova"
#> [115] "MANOVA" "marginaleffects"
#> [117] "marginaleffects.summary" "margins"
#> [119] "maxLik" "mclogit"
#> [121] "mcmc" "mcmc.list"
#> [123] "MCMCglmm" "mcp1"
#> [125] "mcp12" "mcp2"
#> [127] "med1way" "mediate"
#> [129] "merMod" "merModList"
#> [131] "meta_bma" "meta_fixed"
#> [133] "meta_random" "metaplus"
#> [135] "mhurdle" "mipo"
#> [137] "mira" "mixed"
#> [139] "MixMod" "mixor"
#> [141] "mjoint" "mle"
#> [143] "mle2" "mlm"
#> [145] "mlogit" "mmlogit"
#> [147] "model_fit" "multinom"
#> [149] "mvord" "negbinirr"
#> [151] "negbinmfx" "ols"
#> [153] "onesampb" "orm"
#> [155] "pgmm" "plm"
#> [157] "PMCMR" "poissonirr"
#> [159] "poissonmfx" "polr"
#> [161] "probitmfx" "psm"
#> [163] "Rchoice" "ridgelm"
#> [165] "riskRegression" "rjags"
#> [167] "rlm" "rlmerMod"
#> [169] "RM" "rma"
#> [171] "rma.uni" "robmixglm"
#> [173] "robtab" "rq"
#> [175] "rqs" "rqss"
#> [177] "Sarlm" "scam"
#> [179] "selection" "sem"
#> [181] "SemiParBIV" "semLm"
#> [183] "semLme" "slm"
#> [185] "speedglm" "speedlm"
#> [187] "stanfit" "stanmvreg"
#> [189] "stanreg" "summary.lm"
#> [191] "survfit" "survreg"
#> [193] "svy_vglm" "svychisq"
#> [195] "svyglm" "svyolr"
#> [197] "t1way" "tobit"
#> [199] "trimcibt" "truncreg"
#> [201] "vgam" "vglm"
#> [203] "wbgee" "wblm"
#> [205] "wbm" "wmcpAKP"
#> [207] "yuen" "yuend"
#> [209] "zcpglm" "zeroinfl"
#> [211] "zerotrunc"Although not shown here, this function can also be used to carry out parametric, robust, and Bayesian random-effects meta-analysis.
| graphical element | geom_ used |
argument for further modification |
|---|---|---|
| regression estimate | ggplot2::geom_point |
point.args |
| error bars | ggplot2::geom_errorbarh |
errorbar.args |
| vertical line | ggplot2::geom_vline |
vline.args |
| label with statistical details | ggrepel::geom_label_repel |
stats.label.args |
Hypothesis testing and Effect size estimation
| Type | Test | Effect size | CI? | Function used |
|---|---|---|---|---|
| Parametric | Meta-analysis via random-effects models | ✅ | metafor::metafor |
|
| Robust | Meta-analysis via robust random-effects models | ✅ | metaplus::metaplus |
|
| Bayes | Meta-analysis via Bayesian random-effects models | ✅ | metaBMA::meta_random |
For a more exhaustive account of this function, see the associated vignette- https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggcoefstats.html
{ggstatsplot} also offers a convenience function to
extract data frames with statistical details that are used to create
expressions displayed in {ggstatsplot} plots.
set.seed(123)
## a list of tibbles containing statistical analysis summaries
ggbetweenstats(mtcars, cyl, mpg) %>%
extract_stats()
#> $subtitle_data
#> # A tibble: 1 × 14
#> statistic df df.error p.value
#> <dbl> <dbl> <dbl> <dbl>
#> 1 31.6 2 18.0 0.00000127
#> method effectsize estimate
#> <chr> <chr> <dbl>
#> 1 One-way analysis of means (not assuming equal variances) Omega2 0.744
#> conf.level conf.low conf.high conf.method conf.distribution n.obs expression
#> <dbl> <dbl> <dbl> <chr> <chr> <int> <list>
#> 1 0.95 0.531 1 ncp F 32 <language>
#>
#> $caption_data
#> # A tibble: 6 × 18
#> term pd rope.percentage prior.distribution prior.location prior.scale
#> <chr> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 mu 1 0 cauchy 0 0.707
#> 2 cyl-4 1 0 cauchy 0 0.707
#> 3 cyl-6 0.780 0.390 cauchy 0 0.707
#> 4 cyl-8 1 0 cauchy 0 0.707
#> 5 sig2 1 0 cauchy 0 0.707
#> 6 g_cyl 1 0.0155 cauchy 0 0.707
#> bf10 method log_e_bf10 effectsize
#> <dbl> <chr> <dbl> <chr>
#> 1 3008850. Bayes factors for linear models 14.9 Bayesian R-squared
#> 2 3008850. Bayes factors for linear models 14.9 Bayesian R-squared
#> 3 3008850. Bayes factors for linear models 14.9 Bayesian R-squared
#> 4 3008850. Bayes factors for linear models 14.9 Bayesian R-squared
#> 5 3008850. Bayes factors for linear models 14.9 Bayesian R-squared
#> 6 3008850. Bayes factors for linear models 14.9 Bayesian R-squared
#> estimate std.dev conf.level conf.low conf.high conf.method n.obs expression
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <int> <list>
#> 1 0.714 0.0503 0.95 0.574 0.788 HDI 32 <language>
#> 2 0.714 0.0503 0.95 0.574 0.788 HDI 32 <language>
#> 3 0.714 0.0503 0.95 0.574 0.788 HDI 32 <language>
#> 4 0.714 0.0503 0.95 0.574 0.788 HDI 32 <language>
#> 5 0.714 0.0503 0.95 0.574 0.788 HDI 32 <language>
#> 6 0.714 0.0503 0.95 0.574 0.788 HDI 32 <language>
#>
#> $pairwise_comparisons_data
#> # A tibble: 3 × 9
#> group1 group2 statistic p.value alternative distribution p.adjust.method
#> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr>
#> 1 4 6 -6.67 0.00110 two.sided q Holm
#> 2 4 8 -10.7 0.0000140 two.sided q Holm
#> 3 6 8 -7.48 0.000257 two.sided q Holm
#> test expression
#> <chr> <list>
#> 1 Games-Howell <language>
#> 2 Games-Howell <language>
#> 3 Games-Howell <language>
#>
#> $descriptive_data
#> NULL
#>
#> $one_sample_data
#> NULL
#>
#> $tidy_data
#> NULL
#>
#> $glance_data
#> NULLNote that all of this analysis is carried out by
{statsExpressions} package: https://indrajeetpatil.github.io/statsExpressions/
{ggstatsplot} statistical details with custom plotsSometimes you may not like the default plots produced by
{ggstatsplot}. In such cases, you can use other
custom plots (from {ggplot2} or other
plotting packages) and still use {ggstatsplot} functions to
display results from relevant statistical test.
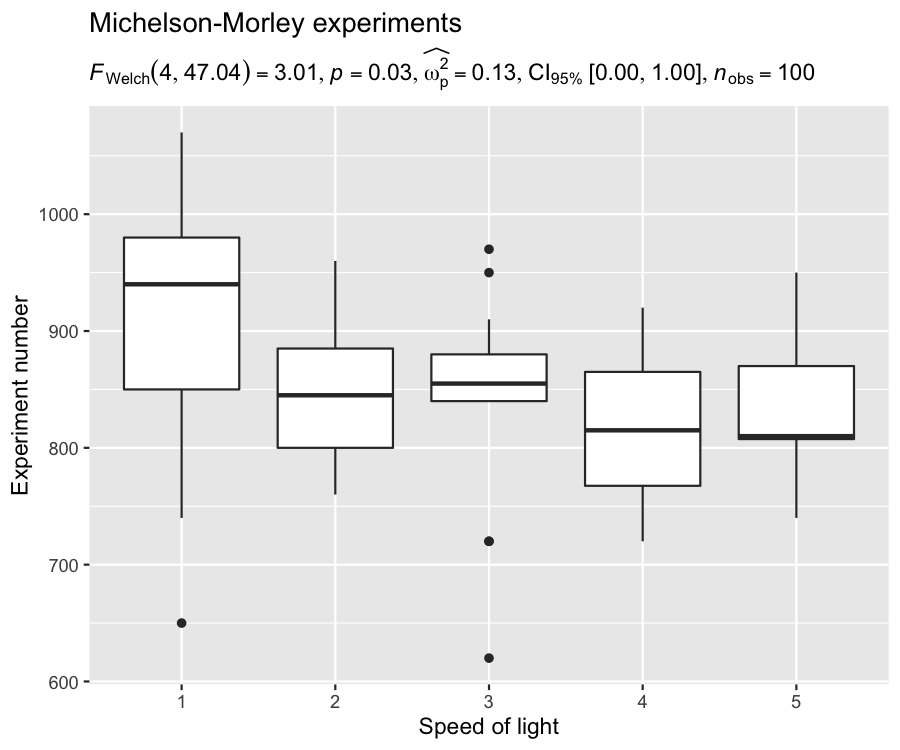
For example, in the following chunk, we will create our own plot
using {ggplot2} package, and use {ggstatsplot}
function for extracting expression:
## loading the needed libraries
set.seed(123)
library(ggplot2)
## using `{ggstatsplot}` to get expression with statistical results
stats_results <- ggbetweenstats(morley, Expt, Speed, output = "subtitle")
## creating a custom plot of our choosing
ggplot(morley, aes(x = as.factor(Expt), y = Speed)) +
geom_boxplot() +
labs(
title = "Michelson-Morley experiments",
subtitle = stats_results,
x = "Speed of light",
y = "Experiment number"
)
{ggstatsplot}No need to use scores of packages for statistical analysis (e.g., one to get stats, one to get effect sizes, another to get Bayes Factors, and yet another to get pairwise comparisons, etc.).
Minimal amount of code needed for all functions (typically only
data, x, and y), which minimizes
chances of error and makes for tidy scripts.
Conveniently toggle between statistical approaches.
Truly makes your figures worth a thousand words.
No need to copy-paste results to the text editor (MS-Word, e.g.).
Disembodied figures stand on their own and are easy to evaluate for the reader.
More breathing room for theoretical discussion and other text.
No need to worry about updating figures and statistical details separately.
{ggstatsplot}This package is…
❌ an alternative to learning {ggplot2}
✅ (The
better you know {ggplot2}, the more you can modify the
defaults to your liking.)
❌ meant to be used in talks/presentations
✅ (Default plots can
be too complicated for effectively communicating results in
time-constrained presentation settings, e.g. conference talks.)
❌ the only game in town
✅ (GUI software alternatives: JASP and jamovi).
In case you use the GUI software jamovi, you can install
a module called jjstatsplot,
which is a wrapper around {ggstatsplot}.
I’m happy to receive bug reports, suggestions, questions, and (most
of all) contributions to fix problems and add features. I personally
prefer using the GitHub issues system over trying to reach
out to me in other ways (personal e-mail, Twitter, etc.). Pull Requests
for contributions are encouraged.
Here are some simple ways in which you can contribute (in the increasing order of commitment):
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.