


mixgb is a scalable multiple imputation framework based
on XGBoost, bootstrapping and predictive mean matching. The proposed
framework is implemented in an R package mixgb. We have
shown that our framework obtains less biased estimates and reflects
appropriate imputation variability, while achieving high computational
efficiency. For more details, please check our paper https://arxiv.org/abs/2106.01574. (Our package has been
revised and updated since the preprint was posted. Simulation code in
the original supplementary files may not run as expected. Revised paper
and adapted code will be updated soon.)
May 2022
April 2022
maxit.m imputations is optional. User
can set bootstrap = FALSE to disable bootstrap. Users can
also set sampling related hyperparameters of XGBoost (subsample,
colsample_bytree, colsample_bylevel, colsample_bynode) to be less than 1
to achieve similar effect.pmm.type are
NULL,0,1,2 or
"auto" (type 2 for numeric/integer variables, no PMM for
categorical variables).data.tablenroundsnthread for
multithreading with OpenMP support. However, MacOS has disabled OpenMP
support.You can install the development version of mixgb from GitHub with:
# install.packages("devtools")
devtools::install_github("agnesdeng/mixgb")# load mixgb
library(mixgb)It is highly recommended to clean and check your data before imputation. Here are some common issues:
NA not
NaNInf or -Inf are not allowedNA or sensible
valuesThe function data_clean() can do a preliminary check and
fix some obvious problems. However, it would not fix all issues related
to data quality.
cleanWithNA.df <- data_clean(rawdata)mixgbWe first load the mixgb package and the
nhanes3_newborn dataset, which contains 16 variables of
various types (integer/numeric/factor/ordinal factor). There are 9
variables with missing values.
str(nhanes3_newborn)
#> tibble [2,107 × 16] (S3: tbl_df/tbl/data.frame)
#> $ HSHSIZER: int [1:2107] 4 3 5 4 4 3 5 3 3 3 ...
#> $ HSAGEIR : int [1:2107] 2 5 10 10 8 3 10 7 2 7 ...
#> $ HSSEX : Factor w/ 2 levels "1","2": 2 1 2 2 1 1 2 2 2 1 ...
#> $ DMARACER: Factor w/ 3 levels "1","2","3": 1 1 2 1 1 1 2 1 2 2 ...
#> $ DMAETHNR: Factor w/ 3 levels "1","2","3": 3 1 3 3 3 3 3 3 3 3 ...
#> $ DMARETHN: Factor w/ 4 levels "1","2","3","4": 1 3 2 1 1 1 2 1 2 2 ...
#> $ BMPHEAD : num [1:2107] 39.3 45.4 43.9 45.8 44.9 42.2 45.8 NA 40.2 44.5 ...
#> ..- attr(*, "label")= chr "Head circumference (cm)"
#> $ BMPRECUM: num [1:2107] 59.5 69.2 69.8 73.8 69 61.7 74.8 NA 64.5 70.2 ...
#> ..- attr(*, "label")= chr "Recumbent length (cm)"
#> $ BMPSB1 : num [1:2107] 8.2 13 6 8 8.2 9.4 5.2 NA 7 5.9 ...
#> ..- attr(*, "label")= chr "First subscapular skinfold (mm)"
#> $ BMPSB2 : num [1:2107] 8 13 5.6 10 7.8 8.4 5.2 NA 7 5.4 ...
#> ..- attr(*, "label")= chr "Second subscapular skinfold (mm)"
#> $ BMPTR1 : num [1:2107] 9 15.6 7 16.4 9.8 9.6 5.8 NA 11 6.8 ...
#> ..- attr(*, "label")= chr "First triceps skinfold (mm)"
#> $ BMPTR2 : num [1:2107] 9.4 14 8.2 12 8.8 8.2 6.6 NA 10.9 7.6 ...
#> ..- attr(*, "label")= chr "Second triceps skinfold (mm)"
#> $ BMPWT : num [1:2107] 6.35 9.45 7.15 10.7 9.35 7.15 8.35 NA 7.35 8.65 ...
#> ..- attr(*, "label")= chr "Weight (kg)"
#> $ DMPPIR : num [1:2107] 3.186 1.269 0.416 2.063 1.464 ...
#> ..- attr(*, "label")= chr "Poverty income ratio"
#> $ HFF1 : Factor w/ 2 levels "1","2": 2 2 1 1 1 2 2 1 2 1 ...
#> $ HYD1 : Ord.factor w/ 5 levels "1"<"2"<"3"<"4"<..: 1 3 1 1 1 1 1 1 2 1 ...
colSums(is.na(nhanes3_newborn))
#> HSHSIZER HSAGEIR HSSEX DMARACER DMAETHNR DMARETHN BMPHEAD BMPRECUM
#> 0 0 0 0 0 0 124 114
#> BMPSB1 BMPSB2 BMPTR1 BMPTR2 BMPWT DMPPIR HFF1 HYD1
#> 161 169 124 167 117 192 7 0To impute this dataset, we can use the default settings. The default
number of imputed datasets m = 5. Note that we do not need
to convert our data into dgCMatrix or one-hot coding format. Our package
will convert it automatically. Variables should be of the following
types: numeric, integer, factor or ordinal factor.
# use mixgb with default settings
imputed.data <- mixgb(data = nhanes3_newborn, m = 5)We can also customise imputation settings:
The number of imputed datasets m
The number of imputation iterations maxit
Whether to convert ordinal factors to integer (imputation process
will be faster) ordinalAsInteger
Whether to use bootstrapping bootstrap
Predictive mean matching settings pmm.type,
pmm.k and pmm.link.
Initial imputation methods for different types of variables
initial.num, initial.int and
initial.fac.
Whether to save models for imputing newdata
save.models and save.vars.
XGBoost hyperparameters and verbose settings.
xgb.params, nrounds,
early_stopping_rounds, print_every_n and
verbose.
# Use mixgb with chosen settings
params <- list(max_depth = 6, gamma = 0.1, eta = 0.3, min_child_weight = 1,
subsample = 1, colsample_bytree = 1, colsample_bylevel = 1,
colsample_bynode = 1, nthread = 4, tree_method = "auto",
gpu_id = 0, predictor = "auto")
imputed.data <- mixgb(data = nhanes3_newborn, m = 5, maxit = 1,
ordinalAsInteger = TRUE, bootstrap = TRUE, pmm.type = "auto",
pmm.k = 5, pmm.link = "prob", initial.num = "normal", initial.int = "mode",
initial.fac = "mode", save.models = FALSE, save.vars = NULL,
xgb.params = params, nrounds = 50, early_stopping_rounds = 10,
print_every_n = 10L, verbose = 0)Imputation performance can be affected by the hyperparameter
settings. It may seem daunting to tune a large set of hyperparameters,
but often we can narrow down the search as many hyperparameters are
correlated. In our package, we have a function mixgb_cv()
to tune nrounds. There is no default nrounds
value in XGBoost, so we need to specify it. The default
nrounds in mixgb is 50. However, we recommend
using mixgb_cv() to find the optimal nrounds
first.
cv.results <- mixgb_cv(data = nhanes3_newborn, verbose = FALSE)
cv.results$response
#> [1] "BMPWT"
cv.results$best.nrounds
#> [1] 18By default, mixgb_cv() will randomly choose an
incomplete variable as the response and build an XGBoost model with
other variables using the complete cases of the dataset. Therefore, each
run of mixgb_cv() is likely to return different results.
Users can also specify the response and covariates in the argument
response and select_features,
respectively.
cv.results <- mixgb_cv(data = nhanes3_newborn, nfold = 10, nrounds = 100,
early_stopping_rounds = 1, response = "BMPHEAD", select_features = c("HSAGEIR",
"HSSEX", "DMARETHN", "BMPRECUM", "BMPSB1", "BMPSB2",
"BMPTR1", "BMPTR2", "BMPWT"), verbose = FALSE)
cv.results$best.nrounds
#> [1] 18Since using mixgb_cv() with this dataset mostly returns
a number less than 20, I’ll set nrounds = 20
in mixgb() to obtain m imputed datasets.
imputed.data <- mixgb(data = nhanes3_newborn, m = 5, nrounds = 20)It is important to assess the plausibility of imputations before
doing analysis. The mixgb package provides several visual
diagnostic functions using ggplot2 to compare multiply
imputed values versus observed data.
We will demonstrate these functions using the
nhanes3_newborn dataset. In the original data, almost all
missing values occurred in numeric variables. Only seven observations
are missing in the binary factor variable HFF1 . In order
to visualize some imputed values for other types of variables, we create
some extra missing values in HSHSIZER (integer),
HSAGEIR (integer), HSSEX (binary factor),
DMARETHN (multiclass factor) and HYD1 (Ordinal
factor) under MCAR.
withNA.df <- createNA(data = nhanes3_newborn, var.names = c("HSHSIZER",
"HSAGEIR", "HSSEX", "DMARETHN", "HYD1"), p = 0.1)
colSums(is.na(withNA.df))
#> HSHSIZER HSAGEIR HSSEX DMARACER DMAETHNR DMARETHN BMPHEAD BMPRECUM
#> 211 211 211 0 0 211 124 114
#> BMPSB1 BMPSB2 BMPTR1 BMPTR2 BMPWT DMPPIR HFF1 HYD1
#> 161 169 124 167 117 192 7 211We then impute this dataset using mixgb() with default
settings. A list of five imputed datasets are assigned to
imputed.data. The dimension of each imputed dataset will be
the same as the original data.
imputed.data <- mixgb(data = withNA.df, m = 5)The mixgb package provides the following visual
diagnostics functions:
Single variable: plot_hist(),
plot_box(), plot_bar() ;
Two variables: plot_2num(),
plot_2fac(), plot_1num1fac() ;
Three variables: plot_2num1fac(),
plot_1num2fac().
Each function will return m+1 panels to compare the
observed data with m sets of actual imputed values.
Here are some examples. For more details, please check the vignette Visual diagnostics for multiply imputed values.
plot_hist(imputation.list = imputed.data, var.name = "BMPHEAD",
original.data = withNA.df)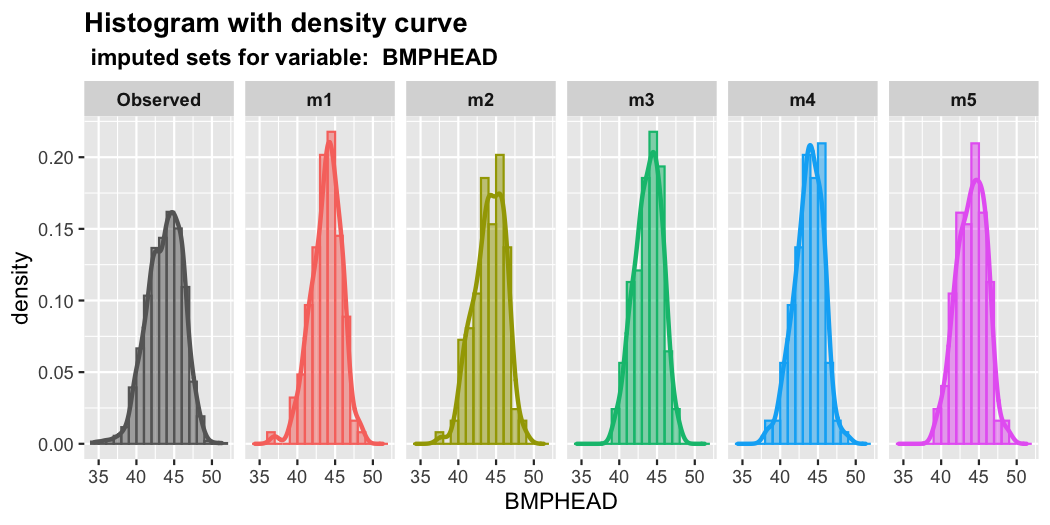
plot_2num(imputation.list = imputed.data, var.x = "BMPHEAD",
var.y = "BMPRECUM", original.data = withNA.df)
plot_2num(imputation.list = imputed.data, var.x = "HSAGEIR",
var.y = "BMPHEAD", original.data = withNA.df)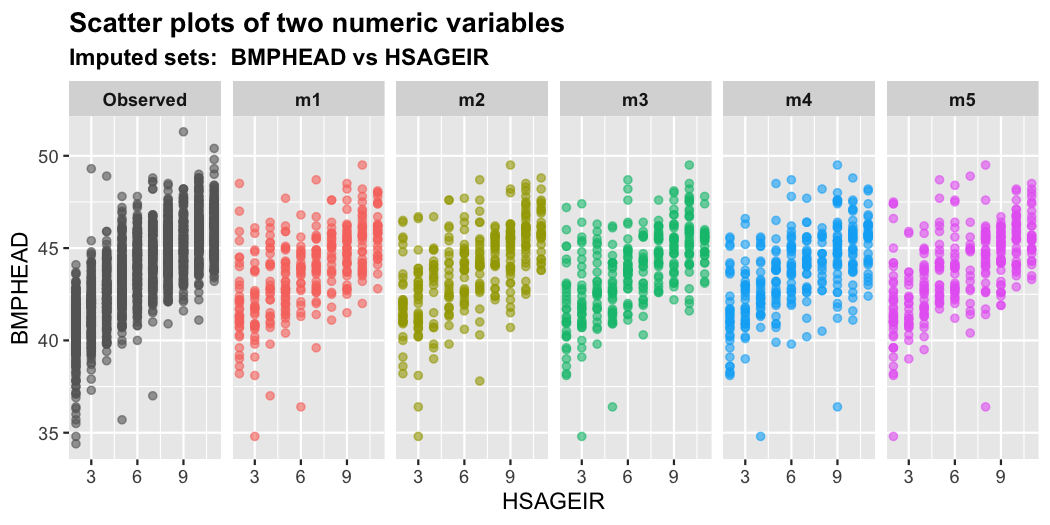
plot_1num1fac(imputation.list = imputed.data, var.num = "BMPHEAD",
var.fac = "HSSEX", original.data = withNA.df)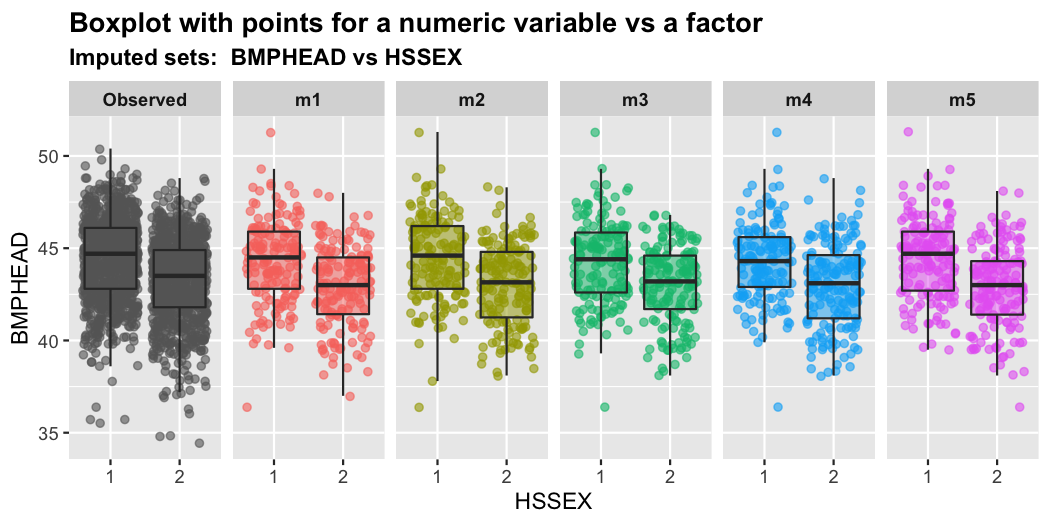
First we can split the nhanes3_newborn dataset into
training data and test data.
library(mixgb)
data("nhanes3_newborn")
set.seed(2022)
n <- nrow(nhanes3_newborn)
idx <- sample(1:n, size = round(0.7 * n), replace = FALSE)
train.data <- nhanes3_newborn[idx, ]
test.data <- nhanes3_newborn[-idx, ]We can use the training data to obtain m imputed
datasets and save their imputation models. To achieve this, users need
to set save.models = TRUE. By default
save.vars = NULL, imputation models for variables with
missing data in the training data will be saved. However, the unseen
data may also have missing values in other variables. Users can be
comprehensive by saving models for all variables by setting
save.vars = colnames(train.data). Note that this would take
much longer as we need to train and save a model for each variable. If
users are confident that only certain variables will have missing values
in the new data, we recommend specifying the names or indices of these
variables in save.vars instead of saving models for all
variables.
# obtain m imputed datasets for train.data and save
# imputation models
mixgb.obj <- mixgb(data = train.data, m = 5, save.models = TRUE,
save.vars = NULL)When save.models = TRUE, mixgb() will
return an object containing the following:
imputed.data: a list of m imputed
dataset for training data
XGB.models: a list of m sets of XGBoost
models for variables specified in save.vars.
params: a list of parameters that are required for
imputing new data using impute_new() later on.
We can extract m imputed datasets from the saved imputer
object by $imputed.data.
train.imputed <- mixgb.obj$imputed.data
# the 5th imputed dataset
head(train.imputed[[5]])
#> HSHSIZER HSAGEIR HSSEX DMARACER DMAETHNR DMARETHN BMPHEAD BMPRECUM BMPSB1
#> 1: 7 2 1 1 1 3 43.0 67.1 9.2
#> 2: 4 3 2 2 3 2 42.6 67.1 8.8
#> 3: 3 9 2 2 3 2 46.5 64.3 8.6
#> 4: 3 9 2 1 3 1 46.2 68.5 10.8
#> 5: 5 4 1 1 3 1 44.7 63.0 6.0
#> 6: 5 10 1 1 3 1 45.2 72.0 5.4
#> BMPSB2 BMPTR1 BMPTR2 BMPWT DMPPIR HFF1 HYD1
#> 1: 8.5 8.8 8.8 7.80 1.701 2 1
#> 2: 8.8 13.3 12.2 8.70 0.102 2 1
#> 3: 8.0 10.4 9.2 8.00 0.359 1 3
#> 4: 10.0 16.6 16.0 8.98 0.561 1 3
#> 5: 5.8 9.0 9.0 7.60 2.379 2 1
#> 6: 5.4 9.2 9.4 9.00 2.173 2 2To impute new data with this saved imputer object, we use the
impute_new() function. User can also specify whether to use
new data for initial imputation. By default,
initial.newdata = FALSE, we will use the information of
training data to initially impute the new data. New data will be imputed
with the saved models. This process will be considerably faster as we
don’t need to build the imputation models again.
test.imputed <- impute_new(object = mixgb.obj, newdata = test.data)If PMM is used when we call mixgb(), predicted values of
missing entries in the new dataset are matched with donors from training
data. Users can also set the number of donors for PMM when imputing new
data. By default, pmm.k = NULL , which means the same
setting as the training object will be used.
Similarly, users can set the number of imputed datasets
m. Note that this value has to be smaller than or equal to
the m in mixgb(). If it is not specified, it
will use the same m value as the saved object.
test.imputed <- impute_new(object = mixgb.obj, newdata = test.data,
initial.newdata = FALSE, pmm.k = 3, m = 4)mixgb
with GPU supportMultiple imputation can be run with GPU support for machines with
NVIDIA GPUs. Note that users have to install the R package
xgboost with GPU support first.
The xgboost R package pre-built binary on Linux x86_64
with GPU support can be downloaded from the release page https://github.com/dmlc/xgboost/releases/tag/v1.4.0
The package can then be installed by running the following commands:
# Install dependencies
$ R -q -e "install.packages(c('data.table', 'jsonlite'))"
# Install XGBoost
$ R CMD INSTALL ./xgboost_r_gpu_linux.tar.gzThen users can install package mixgb in R.
devtools::install_github("agnesdeng/mixgb")
library(mixgb)Users just need to specify tree_method = "gpu_list" in
the params list which will then be passed to xgb.params in
mixgb(). Other GPU-realted arguments include
gpu_id and predictor. By default,
gpu_id = 0 and predictor = "auto".
params <- list(max_depth = 6, gamma = 0.1, eta = 0.3, min_child_weight = 1,
subsample = 1, colsample_bytree = 1, colsample_bylevel = 1,
colsample_bynode = 1, nthread = 4, tree_method = "gpu_list",
gpu_id = 0, predictor = "auto")
mixgb.data <- mixgb(data = withNA.df, m = 5, xgb.params = params)