proxyC computes proximity between rows or columns of large matrices efficiently in C++. It is optimized for large sparse matrices using the Armadillo and Intel TBB libraries. Among several built-in similarity/distance measures, computation of correlation, cosine similarity and Euclidean distance is particularly fast.
This code was originally written for quanteda to compute similarity/distance between documents or features in large corpora, but separated as a stand-alone package to make it available for broader data scientific purposes.
install.packages("proxyC")require(Matrix)
## Loading required package: Matrix
require(microbenchmark)
## Loading required package: microbenchmark
require(RcppParallel)
## Loading required package: RcppParallel
require(ggplot2)
## Loading required package: ggplot2
require(magrittr)
## Loading required package: magrittr
# Set number of threads
setThreadOptions(8)
# Make a matrix with 99% zeros
sm1k <- rsparsematrix(1000, 1000, 0.01) # 1,000 columns
sm10k <- rsparsematrix(1000, 10000, 0.01) # 10,000 columns
# Convert to dense format
dm1k <- as.matrix(sm1k)
dm10k <- as.matrix(sm10k)With sparse matrices, proxyC is roughly 10 to 100 times faster than proxy.
bm1 <- microbenchmark(
"proxy 1k" = proxy::simil(dm1k, method = "cosine"),
"proxyC 1k" = proxyC::simil(sm1k, margin = 2, method = "cosine"),
"proxy 10k" = proxy::simil(dm10k, method = "cosine"),
"proxyC 10k" = proxyC::simil(sm10k, margin = 2, method = "cosine"),
times = 10
)
autoplot(bm1)
## Coordinate system already present. Adding new coordinate system, which will replace the existing one.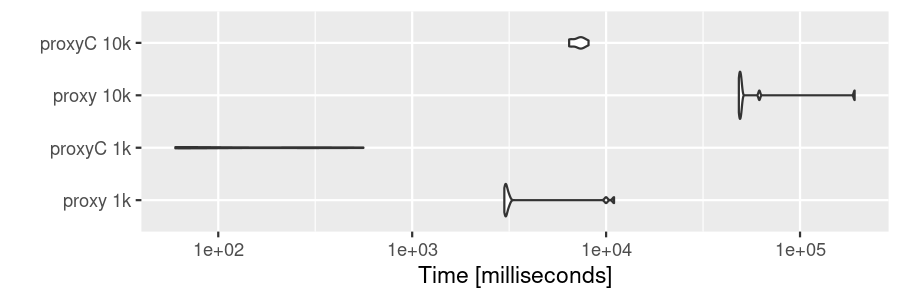
If min_simil is used, proxyC becomes
even faster because small similarity scores are floored to zero.
bm2 <- microbenchmark(
"proxyC all" = proxyC::simil(sm1k, margin = 2, method = "cosine"),
"proxyC min_simil" = proxyC::simil(sm1k, margin = 2, method = "cosine", min_simil = 0.9),
times = 10
)
autoplot(bm2)
## Coordinate system already present. Adding new coordinate system, which will replace the existing one.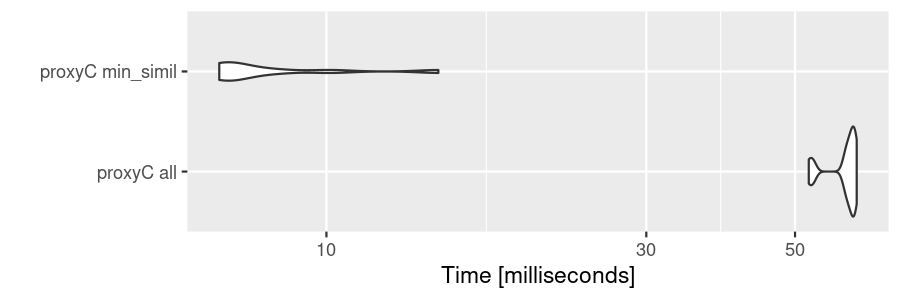
Flooring by min_simil makes the resulting object much
smaller.
proxyC::simil(sm10k, margin = 2, method = "cosine") %>%
object.size() %>%
print(units = "MB")
## 763 Mb
proxyC::simil(sm10k, margin = 2, method = "cosine", min_simil = 0.9) %>%
object.size() %>%
print(units = "MB")
## 0.2 MbIf rank is used, proxyC only returns
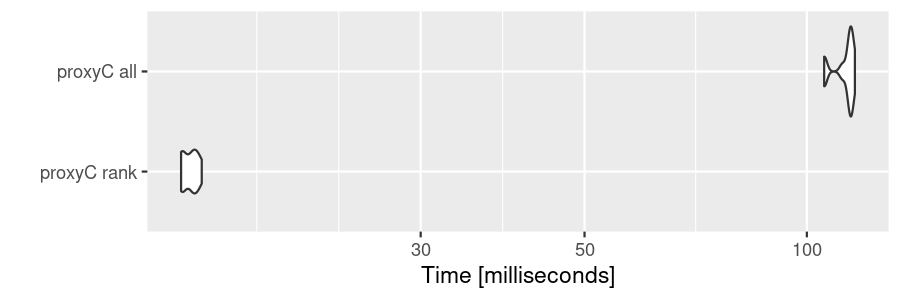
top-n values.
bm3 <- microbenchmark(
"proxyC rank" = proxyC::simil(sm1k, margin = 2, method = "correlation", rank = 10),
"proxyC all" = proxyC::simil(sm1k, margin = 2, method = "correlation"),
times = 10
)
autoplot(bm3)
## Coordinate system already present. Adding new coordinate system, which will replace the existing one.