

NOTE:
restezis no longer available on CRAN due to the archiving of a key dependency (MonetDBLite). It can still be installed via GitHub. The issue is being dealt with and hopefully a new version ofrestezwill be available on CRAN soon.
ADDITIONAL NOTE (2022-07-07): MonetDBLite has now been replaced with duckdb in version 2.0.0, which should allow for submission to CRAN. Because of this change, restez v2.0.0 or higher is not compatible with databases built with previous versions of restez.
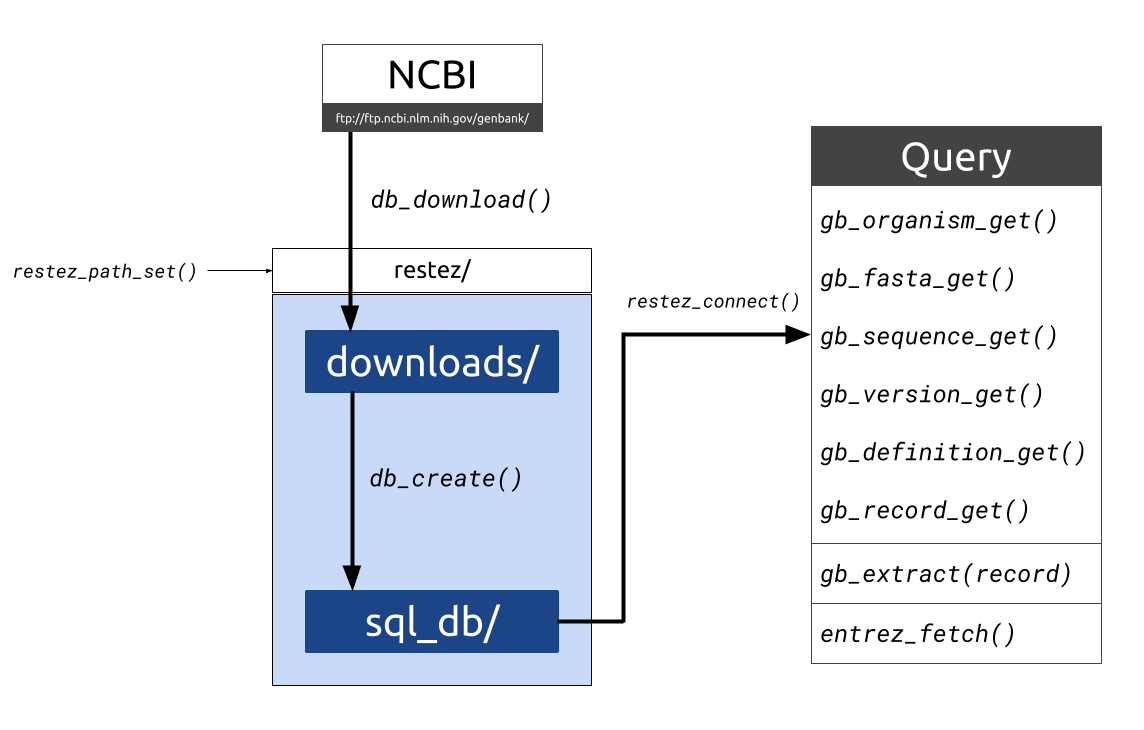
Download parts of NCBI’s GenBank to a local folder and create a simple SQL-like database. Use ‘get’ tools to query the database by accession IDs. rentrez wrappers are available, so that if sequences are not available locally they can be searched for online through Entrez.
See the detailed tutorials for more information.
Vous entrez, vous rentrez et, maintenant, vous …. restez!
Downloading sequences and sequence information from GenBank and related NCBI taxonomic databases is often performed via the NCBI API, Entrez. Entrez, however, has a limit on the number of requests and downloading large amounts of sequence data in this way can be inefficient. For programmatic situations where multiple Entrez calls are made, downloading may take days, weeks or even months.
This package aims to make sequence retrieval more efficient by allowing a user to download large sections of the GenBank database to their local machine and query this local database either through package specific functions or Entrez wrappers. This process is more efficient as GenBank downloads are made via NCBI’s FTP using compressed sequence files. With a good internet connection and a middle-of-the-road computer, a database comprising 20 GB of sequence information can be generated in less than 10 minutes.
The package can currently only be installed through GitHub:
# install.packages("remotes")
remotes::install_github("ropensci/restez")(It was previously available via CRAN but was archived due to a key dependency MonetDBLite being no longer available.)
For more detailed information on the package’s functions and detailed guides on downloading, constructing and querying a database, see the detailed tutorials.
# Warning: running these examples may take a few minutes
library(restez)
# choose a location to store GenBank files
restez_path_set(rstz_pth)# Run the download function
db_download()
# after download, create the local database
db_create()# get a random accession ID from the database
id <- sample(list_db_ids(), 1)
#> Warning in list_db_ids(): Number of ids returned was limited to [100].
#> Set `n=NULL` to return all ids.
# you can extract:
# sequences
seq <- gb_sequence_get(id)[[1]]
str(seq)
#> chr "ACTACTAACTTCAGCCTATCTAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCT"| __truncated__
# definitions
def <- gb_definition_get(id)[[1]]
print(def)
#> [1] "Unidentified sequence clone 1 amplified using OIE white spot virus primers"
# organisms
org <- gb_organism_get(id)[[1]]
print(org)
#> [1] "unidentified"
# or whole records
rec <- gb_record_get(id)[[1]]
cat(rec)
#> LOCUS AY703870 952 bp DNA linear UNA 21-JAN-2005
#> DEFINITION Unidentified sequence clone 1 amplified using OIE white spot virus
#> primers.
#> ACCESSION AY703870
#> VERSION AY703870.1
#> KEYWORDS .
#> SOURCE unidentified
#> ORGANISM unidentified
#> unclassified sequences.
#> REFERENCE 1 (bases 1 to 952)
#> AUTHORS Claydon,K., Cullen,B. and Owens,L.
#> TITLE OIE white spot syndrome virus PCR gives false-positive results in
#> Cherax quadricarinatus
#> JOURNAL Dis. Aquat. Org. 62 (3), 265-268 (2004)
#> PUBMED 15672884
#> REFERENCE 2 (bases 1 to 952)
#> AUTHORS Claydon,K., Cullen,B. and Owens,L.
#> TITLE Direct Submission
#> JOURNAL Submitted (01-AUG-2004) Biomedical Sciences, James Cook University,
#> Solander Drive, Townsville, Qld 4811, Australia
#> FEATURES Location/Qualifiers
#> source 1..952
#> /organism="unidentified"
#> /mol_type="genomic DNA"
#> /db_xref="taxon:32644"
#> /clone="1"
#> misc_feature 1..952
#> /note="false-positive sequence amplified using OIE white
#> spot virus primers"
#> ORIGIN
#> 1 actactaact tcagcctatc tagcactaca ccttcaacat ctccagcact acaccttcaa
#> 61 catctccagc actacacctt caacatctcc agcactacac cttcaacatc tccagcacta
#> 121 caccttcaac atctccagca ctacaccttc aacatctcca gcactacacc ttcagcatct
#> 181 ccagcactac accttcaaca tctccagcac tacatcttca acatctccag cactacacct
#> 241 tcaacatctc cagcactaca ccttcaacat ctccagcact acaccttcaa catctccagc
#> 301 actacacctt caacatctcc agcactacac cttcaacatc tccagcatta cacctttagc
#> 361 atcaccacca ccaccttcaa catcaccacc actacctgca gcactctacc ttcatctaca
#> 421 gtcaagaccc ccaacgtccg ttaagaccaa tgccaagaag ctagatgttt ccaacttgaa
#> 481 ggaattctcc tctagtttgt tcccttgtag tatttctcta atggggctgg ggaagaggta
#> 541 gagggaaggt agagaggaaa aggataaggg agagagggaa agatggaaga ggagaggaag
#> 601 aacgacagaa gaatggattt atgaagaaga gaaggagtag taggaggaga aagagtacgt
#> 661 aagtgcacat gggaagcaga cagcagcaga cattactgtc tacaaggaag ctatctgggg
#> 721 agcactaaca atagtccagc taactactcc ttactggata aatgaccata atttttaaag
#> 781 gggtggaccg gtaagccagc ggaaggcctc ggtcagatga ccaaaagctc caaaggcggg
#> 841 tcatcatctg actaagaccc gcgtcaggaa acatttatcc tgtttcctga cgaaccttac
#> 901 ctaacctaac ctaacctcct tactggaatc tagataggat gaagttagta gt
#> //# use the entrez_* wrappers to access GB data
res <- entrez_fetch(db = 'nucleotide', id = id, rettype = 'fasta')
cat(res)
#> >AY703870.1 Unidentified sequence clone 1 amplified using OIE white spot virus primers
#> ACTACTAACTTCAGCCTATCTAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGC
#> ACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCA
#> CTACACCTTCAACATCTCCAGCACTACACCTTCAGCATCTCCAGCACTACACCTTCAACATCTCCAGCAC
#> TACATCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACT
#> ACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCATTA
#> CACCTTTAGCATCACCACCACCACCTTCAACATCACCACCACTACCTGCAGCACTCTACCTTCATCTACA
#> GTCAAGACCCCCAACGTCCGTTAAGACCAATGCCAAGAAGCTAGATGTTTCCAACTTGAAGGAATTCTCC
#> TCTAGTTTGTTCCCTTGTAGTATTTCTCTAATGGGGCTGGGGAAGAGGTAGAGGGAAGGTAGAGAGGAAA
#> AGGATAAGGGAGAGAGGGAAAGATGGAAGAGGAGAGGAAGAACGACAGAAGAATGGATTTATGAAGAAGA
#> GAAGGAGTAGTAGGAGGAGAAAGAGTACGTAAGTGCACATGGGAAGCAGACAGCAGCAGACATTACTGTC
#> TACAAGGAAGCTATCTGGGGAGCACTAACAATAGTCCAGCTAACTACTCCTTACTGGATAAATGACCATA
#> ATTTTTAAAGGGGTGGACCGGTAAGCCAGCGGAAGGCCTCGGTCAGATGACCAAAAGCTCCAAAGGCGGG
#> TCATCATCTGACTAAGACCCGCGTCAGGAAACATTTATCCTGTTTCCTGACGAACCTTACCTAACCTAAC
#> CTAACCTCCTTACTGGAATCTAGATAGGATGAAGTTAGTAGT
# if the id is not in the local database
# these wrappers will search online via the rentrez package
res <- entrez_fetch(db = 'nucleotide', id = c('S71333.1', id),
rettype = 'fasta')
#> [1] id(s) are unavailable locally, searching online.
cat(res)
#> >AY703870.1 Unidentified sequence clone 1 amplified using OIE white spot virus primers
#> ACTACTAACTTCAGCCTATCTAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGC
#> ACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCA
#> CTACACCTTCAACATCTCCAGCACTACACCTTCAGCATCTCCAGCACTACACCTTCAACATCTCCAGCAC
#> TACATCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACT
#> ACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCACTACACCTTCAACATCTCCAGCATTA
#> CACCTTTAGCATCACCACCACCACCTTCAACATCACCACCACTACCTGCAGCACTCTACCTTCATCTACA
#> GTCAAGACCCCCAACGTCCGTTAAGACCAATGCCAAGAAGCTAGATGTTTCCAACTTGAAGGAATTCTCC
#> TCTAGTTTGTTCCCTTGTAGTATTTCTCTAATGGGGCTGGGGAAGAGGTAGAGGGAAGGTAGAGAGGAAA
#> AGGATAAGGGAGAGAGGGAAAGATGGAAGAGGAGAGGAAGAACGACAGAAGAATGGATTTATGAAGAAGA
#> GAAGGAGTAGTAGGAGGAGAAAGAGTACGTAAGTGCACATGGGAAGCAGACAGCAGCAGACATTACTGTC
#> TACAAGGAAGCTATCTGGGGAGCACTAACAATAGTCCAGCTAACTACTCCTTACTGGATAAATGACCATA
#> ATTTTTAAAGGGGTGGACCGGTAAGCCAGCGGAAGGCCTCGGTCAGATGACCAAAAGCTCCAAAGGCGGG
#> TCATCATCTGACTAAGACCCGCGTCAGGAAACATTTATCCTGTTTCCTGACGAACCTTACCTAACCTAAC
#> CTAACCTCCTTACTGGAATCTAGATAGGATGAAGTTAGTAGT
#>
#> >S71333.1 alpha 1,3 galactosyltransferase [New World monkeys, mermoset lymphoid cell line B95.8, mRNA Partial, 1131 nt]
#> ATGAATGTCAAAGGAAAAGTAATTCTGTCGATGCTGGTTGTCTCAACTGTGATTGTTGTGTTTTGGGAAT
#> ATATCAACAGCCCAGAAGGCTCTTTCTTGTGGATATATCACTCAAAGAACCCAGAAGTTGATGACAGCAG
#> TGCTCAGAAGGACTGGTGGTTTCCTGGCTGGTTTAACAATGGGATCCACAATTATCAACAAGAGGAAGAA
#> GACACAGACAAAGAAAAAGGAAGAGAGGAGGAACAAAAAAAGGAAGATGACACAACAGAGCTTCGGCTAT
#> GGGACTGGTTTAATCCAAAGAAACGCCCAGAGGTTATGACAGTGACCCAATGGAAGGCGCCGGTTGTGTG
#> GGAAGGCACTTACAACAAAGCCATCCTAGAAAATTATTATGCCAAACAGAAAATTACCGTGGGGTTGACG
#> GTTTTTGCTATTGGAAGATATATTGAGCATTACTTGGAGGAGTTCGTAACATCTGCTAATAGGTACTTCA
#> TGGTCGGCCACAAAGTCATATTTTATGTCATGGTGGATGATGTCTCCAAGGCGCCGTTTATAGAGCTGGG
#> TCCTCTGCGTTCCTTCAAAGTGTTTGAGGTCAAGCCAGAGAAGAGGTGGCAAGACATCAGCATGATGCGT
#> ATGAAGACCATCGGGGAGCACATCTTGGCCCACATCCAACACGAGGTTGACTTCCTCTTCTGCATGGATG
#> TGGACCAGGTCTTCCAAGACCATTTTGGGGTAGAGACCCTGGGCCAGTCGGTGGCTCAGCTACAGGCCTG
#> GTGGTACAAGGCAGATCCTGATGACTTTACCTATGAGAGGCGGAAAGAGTCGGCAGCATATATTCCATTT
#> GGCCAGGGGGATTTTTATTACCATGCAGCCATTTTTGGAGGAACACCGATTCAGGTTCTCAACATCACCC
#> AGGAGTGCTTTAAGGGAATCCTCCTGGACAAGAAAAATGACATAGAAGCCGAGTGGCATGATGAAAGCCA
#> CCTAAACAAGTATTTCCTTCTCAACAAACCCTCTAAAATCTTATCTCCAGAATACTGCTGGGATTATCAT
#> ATAGGCCTGCCTTCAGATATTAAAACTGTCAAGCTATCATGGCAAACAAAAGAGTATAATTTGGTTAGAA
#> AGAATGTCTGAWant to contribute? Check the contributing page.
MIT
Bennett et al. (2018). restez: Create and Query a Local Copy of GenBank in R. Journal of Open Source Software, 3(31), 1102. https://doi.org/10.21105/joss.01102
Benson, D. A., Karsch-Mizrachi, I., Clark, K., Lipman, D. J., Ostell, J., & Sayers, E. W. (2012). GenBank. Nucleic Acids Research, 40(Database issue), D48–D53. http://doi.org/10.1093/nar/gkr1202
Winter DJ. (2017) rentrez: An R package for the NCBI eUtils API. PeerJ Preprints 5:e3179v2 https://doi.org/10.7287/peerj.preprints.3179v2
This package previously developed and maintained by Dom Bennett