
The first R package to work with GEXF graph files (used in Gephi and others). rgexf allows the user to quickly build/read graph files, including:
Nodes/edges attributes,
GEXF viz attributes (such as color, size, and position),
Network dynamics (for both edges and nodes, including spells), and
Edges weighting.
Users can build/handle graphs element-by-element or through data-frames, visualize the graph on a web browser through sigmajs javascript gexf-js library and interact with the igraph package.
We now have a hex sticker!
Users can now cite properly rgexf with JOSS (see citation(package="rgexf")).
Added a few extra breaks across the examples (suggested by @corneliusfritz).
Improved documentation regarding spells and dynamic graphs (suggested by @jonjoncardoso).
New head() function allows a glimpse of the n first nodes/edges.
Passing colors with four values (alpha) no longer fails during checks. (reported by @IsabelFE).
The summary function prints nodes’ attributes as expected.
Hex colors now work (#41 reported by @milnus).
gexf.to.igraph correctly processes edge attributes (#38 reported by @balachia).
Time range is now correctly computed (#19).
Non-integer ids were incorrectly processed when reading GEXF files.
More in the NEWS.md file.
To install the latest version of rgexf you can use devtools
The more stable (but old) version of rgexf can be found on CRAN too:
install.packages("rgexf")To cite rgexf in publications use the following paper:
Vega Yon, G. G., (2021). Building, Importing, and Exporting GEXF
Graph Files with rgexf. Journal of Open Source Software, 6(64), 3456,
https://doi.org/10.21105/joss.03456
And the actual R package:
Vega Yon G, Fábrega Lacoa J, Kunst J (2021). _netdiffuseR: Build,
Import and Export GEXF Graph Files_. doi: 10.5281/zenodo.5182708 (URL:
https://doi.org/10.5281/zenodo.5182708), R package version 0.16.2,
<URL: https://github.com/gvegayon/rgexf>.
To see these entries in BibTeX format, use 'print(<citation>,
bibtex=TRUE)', 'toBibtex(.)', or set
'options(citation.bibtex.max=999)'.We can use the read.gexf function to read GEXF files into R:
# Loading the package
library(rgexf)
g <- system.file("gexf-graphs/lesmiserables.gexf", package="rgexf")
g <- read.gexf(g)
head(g) # Taking a look at the first handful## <?xml version="1.0" encoding="UTF-8"?>
## <gexf xmlns="http://www.gexf.net/1.3" xmlns:viz="http://www.gexf.net/1.3/viz" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" version="1.3" xsi:schemaLocation="http://www.gexf.net/1.3 http://www.gexf.net/1.3/gexf.xsd">
## <meta lastmodifieddate="2016-11-09">
## <creator>Gephi 0.9</creator>
## <description/>
## </meta>
## <graph defaultedgetype="undirected" mode="static">
## <attributes class="node" mode="static">
## <attribute id="modularity_class" title="Modularity Class" type="integer"/>
## </attributes>
## <nodes>
## <node id="11" label="Valjean">
## <attvalues>
## <attvalue for="modularity_class" value="1"/>
## </attvalues>
## <viz:size value="100.0"/>
## <viz:position x="-87.93029" y="6.8120565"/>
## <viz:color r="245" g="91" b="91"/>
## </node>
## <node id="48" label="Gavroche">
## <attvalues>
## <attvalue for="modularity_class" value="8"/>
## </attvalues>
## <viz:size value="61.600006"/>
## <viz:position x="387.89572" y="-110.462326"/>
## <viz:color r="91" g="245" b="91"/>
## </node>
## <node id="55" label="Marius">
## <attvalues>
## <attvalue for="modularity_class" value="6"/>
## </attvalues>
## <viz:size value="53.37143"/>
## <viz:position x="206.44687" y="13.805411"/>
## <viz:color r="194" g="91" b="245"/>
## </node>
## <node id="27" label="Javert">
## <attvalues>
## <attvalue for="modularity_class" value="7"/>
## </attvalues>
## <viz:size value="47.88571"/>
## <viz:position x="-81.46074" y="204.20204"/>
## <viz:color r="91" g="245" b="194"/>
## </node>
## <node id="25" label="Thenardier">
## <attvalues>
## <attvalue for="modularity_class" value="7"/>
## </attvalues>
## <viz:size value="45.142853"/>
## <viz:position x="82.80825" y="203.1144"/>
## <viz:color r="91" g="245" b="194"/>
## </node>
## <node id="23" label="Fantine">
## <attvalues>
## <attvalue for="modularity_class" value="2"/>
## </attvalues>
## <viz:size value="42.4"/>
## <viz:position x="-313.42786" y="289.44803"/>
## <viz:color r="91" g="194" b="245"/>
## </node>
## ...
## </nodes>
## <edges>
## <edge id="0" source="1" target="0"/>
## <edge id="1" source="2" target="0" weight="8.0"/>
## <edge id="2" source="3" target="0" weight="10.0"/>
## <edge id="3" source="3" target="2" weight="6.0"/>
## <edge id="4" source="4" target="0"/>
## <edge id="5" source="5" target="0"/>
## ...
## </edges>
## </graph>
## </gexf>Moreover, we can use the gexf.to.igraph() function to convert the gexf object into an igraph object:
##
## Attaching package: 'igraph'
## The following objects are masked from 'package:stats':
##
## decompose, spectrum
## The following object is masked from 'package:base':
##
## union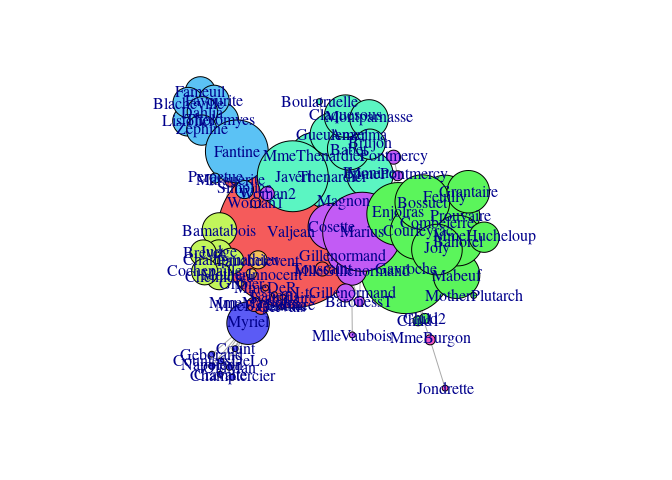
Using the plot.gexf method–which uses the gexf-js JavaScript library–results in a Web visualization of the graph, like this:
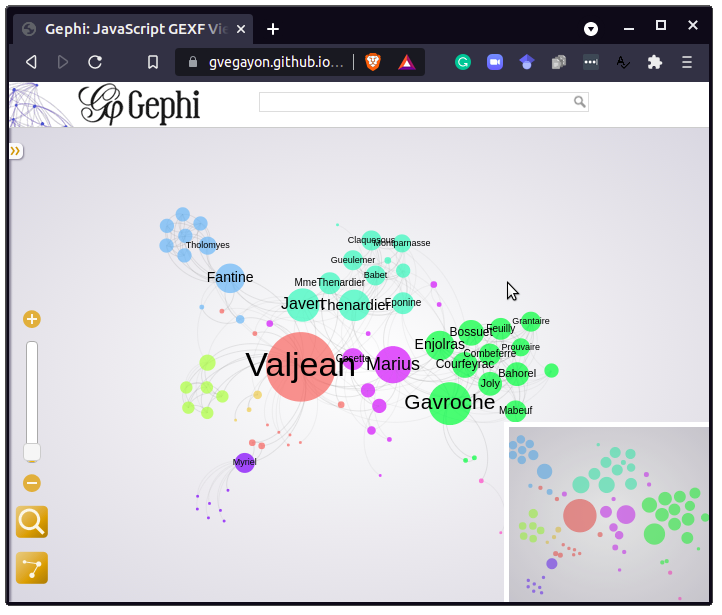
An live version of the figure is available here.
# Creating a group of individuals and their relations
people <- data.frame(matrix(c(1:4, 'juan', 'pedro', 'matthew', 'carlos'),ncol=2))
people## X1 X2
## 1 1 juan
## 2 2 pedro
## 3 3 matthew
## 4 4 carlos# Defining the relations structure
relations <- data.frame(matrix(c(1,4,1,2,1,3,2,3,3,4,4,2), ncol=2, byrow=T))
relations## X1 X2
## 1 1 4
## 2 1 2
## 3 1 3
## 4 2 3
## 5 3 4
## 6 4 2## <?xml version="1.0" encoding="UTF-8"?>
## <gexf xmlns="http://www.gexf.net/1.3" xmlns:viz="http://www.gexf.net/1.3/viz" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.gexf.net/1.3 http://www.gexf.net/1.3/gexf.xsd" version="1.3">
## <meta lastmodifieddate="2021-08-12">
## <creator>NodosChile</creator>
## <description>A GEXF file written in R with "rgexf"</description>
## <keywords>GEXF, NodosChile, R, rgexf, Gephi</keywords>
## </meta>
## <graph mode="static" defaultedgetype="undirected">
## <nodes>
## <node id="1" label="juan">
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="100" y="76.2515713120152" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="2" label="pedro">
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="-100" y="-100" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="3" label="matthew">
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="-33.934012157033" y="55.4559262362359" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="4" label="carlos">
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="4.50330959513599" y="100" z="0"/>
## <viz:size value="10"/>
## </node>
## </nodes>
## <edges>
## <edge id="0" source="1" target="4" weight="1"/>
## <edge id="1" source="1" target="2" weight="1"/>
## <edge id="2" source="1" target="3" weight="1"/>
## <edge id="3" source="2" target="3" weight="1"/>
## <edge id="4" source="3" target="4" weight="1"/>
## <edge id="5" source="4" target="2" weight="1"/>
## </edges>
## </graph>
## </gexf>
## # Defining the dynamic structure, note that there are some nodes that have NA at the end.
time<-matrix(c(10.0,13.0,2.0,2.0,12.0,rep(NA,3)), nrow=4, ncol=2)
time## [,1] [,2]
## [1,] 10 12
## [2,] 13 NA
## [3,] 2 NA
## [4,] 2 NA## <?xml version="1.0" encoding="UTF-8"?>
## <gexf xmlns="http://www.gexf.net/1.3" xmlns:viz="http://www.gexf.net/1.3/viz" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.gexf.net/1.3 http://www.gexf.net/1.3/gexf.xsd" version="1.3">
## <meta lastmodifieddate="2021-08-12">
## <creator>NodosChile</creator>
## <description>A GEXF file written in R with "rgexf"</description>
## <keywords>GEXF, NodosChile, R, rgexf, Gephi</keywords>
## </meta>
## <graph mode="dynamic" start="2" end="13" timeformat="double" defaultedgetype="undirected">
## <nodes>
## <node id="1" label="juan" start="10" end="12">
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="93.5382155239999" y="100" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="2" label="pedro" start="13" end="13">
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="-100" y="-45.2089025903595" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="3" label="matthew" start="2" end="13">
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="100" y="-25.9442855933132" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="4" label="carlos" start="2" end="13">
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="-10.7885944593362" y="-100" z="0"/>
## <viz:size value="10"/>
## </node>
## </nodes>
## <edges>
## <edge id="0" source="1" target="4" weight="1"/>
## <edge id="1" source="1" target="2" weight="1"/>
## <edge id="2" source="1" target="3" weight="1"/>
## <edge id="3" source="2" target="3" weight="1"/>
## <edge id="4" source="3" target="4" weight="1"/>
## <edge id="5" source="4" target="2" weight="1"/>
## </edges>
## </graph>
## </gexf>
## First we define dynamics
## [,1] [,2]
## [1,] 10 12
## [2,] 13 NA
## [3,] 2 NA
## [4,] 2 NA## [,1] [,2]
## [1,] 10 5
## [2,] 13 NA
## [3,] 2 NA
## [4,] 2 NA
## [5,] 12 NA
## [6,] 1 NANow we define the attribute values
# Defining a data frame of attributes for nodes and edges
node.att <- data.frame(letrafavorita=letters[1:4], numbers=1:4, stringsAsFactors=F)
node.att## letrafavorita numbers
## 1 a 1
## 2 b 2
## 3 c 3
## 4 d 4## letrafavorita numbers
## 1 a 1
## 2 b 2
## 3 c 3
## 4 d 4
## 5 e 5
## 6 f 6# Getting the things done
write.gexf(nodes=people, edges=relations, edgeDynamic=time.edges,
edgesAtt=edge.att, nodeDynamic=time.nodes, nodesAtt=node.att)## <?xml version="1.0" encoding="UTF-8"?>
## <gexf xmlns="http://www.gexf.net/1.3" xmlns:viz="http://www.gexf.net/1.3/viz" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.gexf.net/1.3 http://www.gexf.net/1.3/gexf.xsd" version="1.3">
## <meta lastmodifieddate="2021-08-12">
## <creator>NodosChile</creator>
## <description>A GEXF file written in R with "rgexf"</description>
## <keywords>GEXF, NodosChile, R, rgexf, Gephi</keywords>
## </meta>
## <graph mode="dynamic" start="1" end="13" timeformat="double" defaultedgetype="undirected">
## <attributes class="node" mode="static">
## <attribute id="att1" title="letrafavorita" type="string"/>
## <attribute id="att2" title="numbers" type="integer"/>
## </attributes>
## <attributes class="edge" mode="static">
## <attribute id="att1" title="letrafavorita" type="string"/>
## <attribute id="att2" title="numbers" type="integer"/>
## </attributes>
## <nodes>
## <node id="1" label="juan" start="10" end="12">
## <attvalues>
## <attvalue for="att1" value="a"/>
## <attvalue for="att2" value="1"/>
## </attvalues>
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="-52.8911775840454" y="-100" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="2" label="pedro" start="13" end="13">
## <attvalues>
## <attvalue for="att1" value="b"/>
## <attvalue for="att2" value="2"/>
## </attvalues>
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="-79.9134848326296" y="-8.92670659506044" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="3" label="matthew" start="2" end="13">
## <attvalues>
## <attvalue for="att1" value="c"/>
## <attvalue for="att2" value="3"/>
## </attvalues>
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="100" y="100" z="0"/>
## <viz:size value="10"/>
## </node>
## <node id="4" label="carlos" start="2" end="13">
## <attvalues>
## <attvalue for="att1" value="d"/>
## <attvalue for="att2" value="4"/>
## </attvalues>
## <viz:color r="255" g="99" b="71" a="1"/>
## <viz:position x="-100" y="-40.0433660159618" z="0"/>
## <viz:size value="10"/>
## </node>
## </nodes>
## <edges>
## <edge id="0" source="1" target="4" start="10" end="5" weight="1">
## <attvalues>
## <attvalue for="att1" value="a"/>
## <attvalue for="att2" value="1"/>
## </attvalues>
## </edge>
## <edge id="1" source="1" target="2" start="13" end="13" weight="1">
## <attvalues>
## <attvalue for="att1" value="b"/>
## <attvalue for="att2" value="2"/>
## </attvalues>
## </edge>
## <edge id="2" source="1" target="3" start="2" end="13" weight="1">
## <attvalues>
## <attvalue for="att1" value="c"/>
## <attvalue for="att2" value="3"/>
## </attvalues>
## </edge>
## <edge id="3" source="2" target="3" start="2" end="13" weight="1">
## <attvalues>
## <attvalue for="att1" value="d"/>
## <attvalue for="att2" value="4"/>
## </attvalues>
## </edge>
## <edge id="4" source="3" target="4" start="12" end="13" weight="1">
## <attvalues>
## <attvalue for="att1" value="e"/>
## <attvalue for="att2" value="5"/>
## </attvalues>
## </edge>
## <edge id="5" source="4" target="2" start="1" end="13" weight="1">
## <attvalues>
## <attvalue for="att1" value="f"/>
## <attvalue for="att2" value="6"/>
## </attvalues>
## </edge>
## </edges>
## </graph>
## </gexf>
## We welcome contributions to rgexf. Whether reporting a bug, starting a discussion by asking a question, or proposing/requesting a new feature, please go by creating a new issue here so that we can talk about it.
Please note that the rgexf project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.0 (2021-05-18)
## os Ubuntu 18.04.5 LTS
## system x86_64, linux-gnu
## ui X11
## language en_US:en
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/Los_Angeles
## date 2021-08-12
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date lib source
## cachem 1.0.5 2021-05-15 [1] CRAN (R 4.1.0)
## callr 3.7.0 2021-04-20 [1] CRAN (R 4.1.0)
## cli 3.0.0 2021-06-30 [1] CRAN (R 4.1.0)
## crayon 1.4.1 2021-02-08 [1] CRAN (R 4.1.0)
## desc 1.3.0 2021-03-05 [1] CRAN (R 4.1.0)
## devtools 2.4.2 2021-06-07 [1] CRAN (R 4.1.0)
## digest 0.6.27 2020-10-24 [1] CRAN (R 4.1.0)
## ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.1.0)
## evaluate 0.14 2019-05-28 [1] CRAN (R 4.1.0)
## fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.0)
## fs 1.5.0 2020-07-31 [1] CRAN (R 4.1.0)
## glue 1.4.2 2020-08-27 [1] CRAN (R 4.1.0)
## highr 0.9 2021-04-16 [1] CRAN (R 4.1.0)
## htmltools 0.5.1.1 2021-01-22 [1] CRAN (R 4.1.0)
## httpuv 1.6.1 2021-05-07 [1] CRAN (R 4.1.0)
## igraph * 1.2.6 2020-10-06 [1] CRAN (R 4.1.0)
## jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.1.0)
## knitr 1.33 2021-04-24 [1] CRAN (R 4.1.0)
## later 1.2.0 2021-04-23 [1] CRAN (R 4.1.0)
## lifecycle 1.0.0 2021-02-15 [1] CRAN (R 4.1.0)
## magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.1.0)
## memoise 2.0.0 2021-01-26 [1] CRAN (R 4.1.0)
## pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.1.0)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.1.0)
## pkgload 1.2.1 2021-04-06 [1] CRAN (R 4.1.0)
## prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.1.0)
## processx 3.5.2 2021-04-30 [1] CRAN (R 4.1.0)
## promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.1.0)
## ps 1.6.0 2021-02-28 [1] CRAN (R 4.1.0)
## purrr 0.3.4 2020-04-17 [1] CRAN (R 4.1.0)
## R6 2.5.0 2020-10-28 [1] CRAN (R 4.1.0)
## Rcpp 1.0.6 2021-01-15 [1] CRAN (R 4.1.0)
## remotes 2.3.0 2021-04-01 [1] CRAN (R 4.1.0)
## rgexf * 0.16.2 2021-08-12 [1] local
## rlang 0.4.11 2021-04-30 [1] CRAN (R 4.1.0)
## rmarkdown 2.8 2021-05-07 [1] CRAN (R 4.1.0)
## rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.1.0)
## servr 0.22 2021-04-14 [1] CRAN (R 4.1.0)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.1.0)
## stringi 1.6.2 2021-05-17 [1] CRAN (R 4.1.0)
## stringr 1.4.0 2019-02-10 [1] CRAN (R 4.1.0)
## testthat 3.0.2 2021-02-14 [1] CRAN (R 4.1.0)
## usethis 2.0.1 2021-02-10 [1] CRAN (R 4.1.0)
## withr 2.4.2 2021-04-18 [1] CRAN (R 4.1.0)
## xfun 0.23 2021-05-15 [1] CRAN (R 4.1.0)
## XML 3.99-0.6 2021-03-16 [1] CRAN (R 4.1.0)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.1.0)
##
## [1] /home/george/R/x86_64-pc-linux-gnu-library/4.1
## [2] /usr/local/lib/R/site-library
## [3] /usr/lib/R/site-library
## [4] /usr/lib/R/library