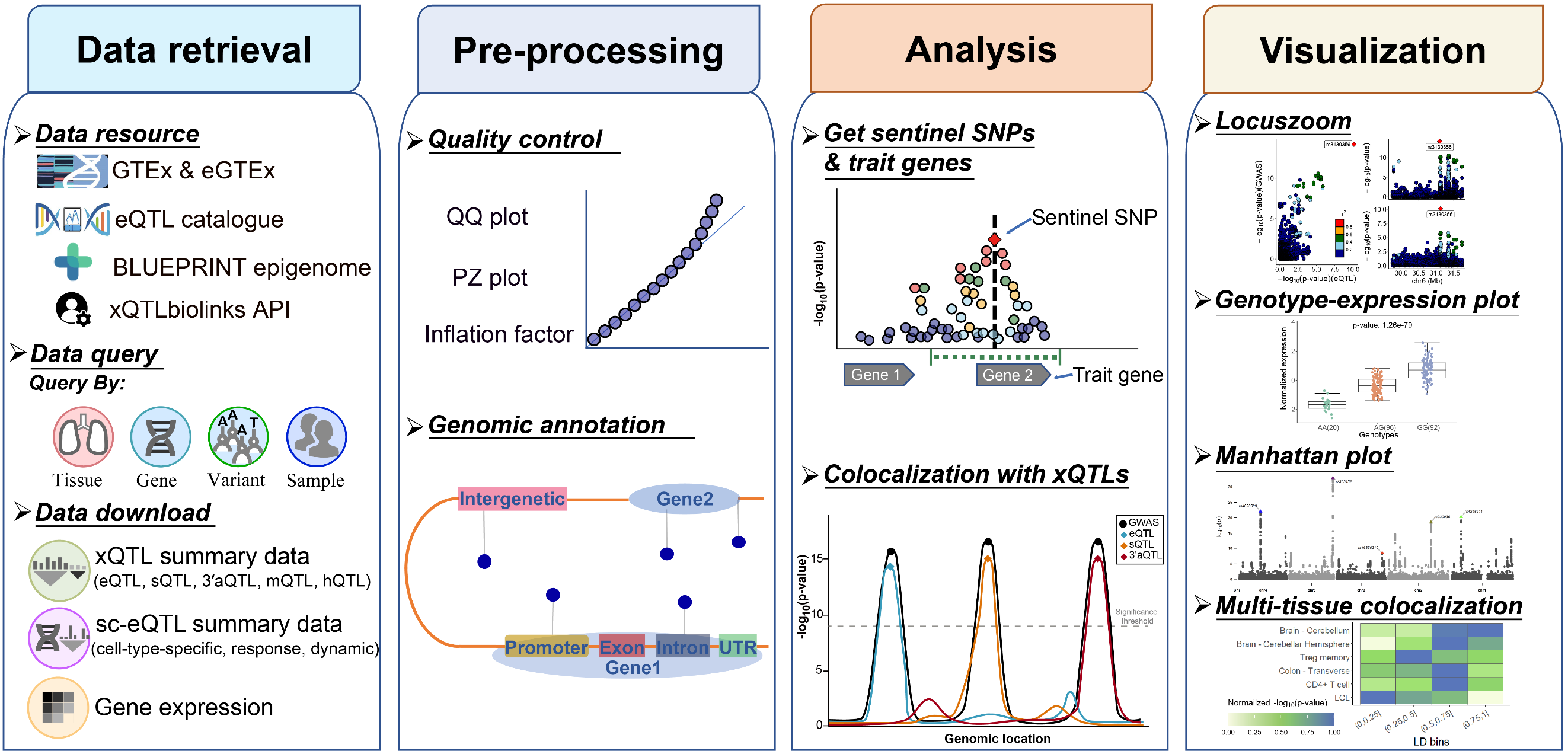
xQTLbiolinks is a well-developed R
package that enables users-customized query, extraction, and
visualization of molecular QTLs (eQTLs and sQTLs) and
gene expression data from public resources (e.g., GTEx)
through the application programming interface (API) of GTEx and eQTL Catalogue.
xQTLbiolinks consists of tailored functions that can be grouped into four modules: Query, Download, Analyze and Visualization.
Instructions, documentation, and tutorials can be found at here.

xQTLbiolinks has been successfully installed on Mac OS
X, Linux, and Windows, using the install.package function
to install directly from CRAN with command
install.package("xQTLbiolinks"). Check more details in
section “Installation” below.xQTLbiolinks.If you find the xQTLbiolinks package or any of the source code in this repository useful for your work, please cite:
Ruofan Ding, Xudong Zou, Yangmei Qin, Gao Wang, Lei Li. xQTLbiolinks: an R package for integrative analysis of xQTL data. (submitted)
Institute of Systems and Physical Biology, Shenzhen Bay Laboratory, Shenzhen 518055, China
We strongly recommend to install in the environment of R, instead of Rstudio.
Package SummarizedExperiment is required but
reposited in Biocouductor. If you haven’t installed it, please install
SummarizedExperiment from Biocouductor with following
command.
# install required bioconductor package SummarizedExperiment:
if (!require("BiocManager")){install.packages("BiocManager")}
BiocManager::install("SummarizedExperiment")xQTLbiolinks from CRAN or github.# This command should automatically install any missing dependencies that are available from CRAN
install.packages("xQTLbiolinks")
# Or install from github.
if(!require("remotes")){install.packages("remotes")}
remotes::install_github("dingruofan/xQTLbiolinks")